Abstract
Cyclin-dependent kinase 7 (Cdk7) is the catalytic subunit of the metazoan Cdk-activating kinase (CAK). Activation of Cdk7 requires its association with a regulatory subunit, Cyclin H. Although the Cdk7/Cyclin H complex has been implicated in the regulation of RNA polymerase in several species, the precise function of their orthologs in zebrafish has not been fully elucidated. In this study, we isolated from zebrafish blastula embryos two cDNAs encoding the orthologs of human Cyclin H and Cdk7, and examined the role of Cdk7/Cyclin H in zebrafish embryogenesis. Sequence analysis showed that the zebrafish Cyclin H and Cdk7 cDNAs encode proteins with 65% and 86% identity to the respective human orthologs. RT-PCR and whole-mount in situ hybridization analyses of their expression in unfertilized eggs, embryos and organs of adult fish suggested that Cyclin H and Cdk7 messages are maternally loaded. Our data also showed that their transcripts were detected throughout development. Distribution of Cyclin H transcripts was found to be ubiquitous during early stages of development and become restricted to the anterior neural tube, brain, eyes, procreate tissues, liver and heart by 5 days post-fertilization. Expression of a dominant-negative form of Cyclin H delayed the onset of zygotic transcription in the early embryo, resulting in apoptosis at 5 hours post-fertilization and leading to sever defects in tissues normally exhibiting high levels of Cyclin H expression. These results implicate Cyclin H in the regulation of the transcriptional machinery during midblastula transition and suggest that it is an essential gene in early zebrafish larval development.
Similar content being viewed by others
Introduction
Metazoan cyclin-dependent kinase 7 (Cdk7), also known as MO15, was initially identified from a search for cDNAs encoding protein kinase(s) related to Cdk1 1. Activation of Cdk1, Cdk2, Cdk4 and Cdk6 by Cdk7 2, the catalytic subunit of the Cdk-activating kinase (CAK), requires the association of Cdk7 with a regulatory subunit, Cyclin H, and the phosphorylation of a conserved threonine residue at position 170 within its own T loop 3. Additionally, CAK is found in the general transcription factor complex TFIIH and phosphorylates the RNA polymerase II carboxyl-terminal domain (CTD) 4. To date, two forms of CAK have been identified: the first contains Cdk7, Cyclin H and an assembly factor known as MAT-1, while the second and minor form lacks MAT-1 5. Previous research has suggested that MAT-1 promoted the assembly of Cdk7 and Cyclin H in vitro, stabilized the transient Cdk7-Cyclin H complex and enhanced specific binding of CAK to TFIIH 6, 7, 8. Collectively, these data suggest a dual role for metazoan Cdk7/Cyclin H both as a CAK essential for cell cycle progression, and as an important CTD kinase.
Although the Cdk7 ortholog in the budding yeast Saccharomyces cerevisiae, encoded by the KIN28 gene, is a strong candidate as the physiological CTD kinase required for mRNA transcription, it has not been demonstrated to regulate the phosphorylation state of the yeast cell cycle Cdk, nor has it been shown to possess CAK activity in vitro 9. The CAK from S. cerevisiae was identified as Cak1, suggesting that budding yeast and vertebrates may have evolved different mechanisms of Cdk activation. The CAK and CTD kinase functions are split in budding yeast, with Kin28p as the kinase subunit of TFIIH and Cak1p as a CAK 10. Similar mechanisms have been identified in S. prombe and Arabidopsis, where multiple CAKs are implicated in the regulation of Cdk and/or CTD phosphorylation 11, 12. Recently, cdk2-affinity chromatography using lysates of HeLa cells identified a novel CAK of approximately 30–40 kDa in human cells 13. Moreover, different CAKs were identified in TGF-beta-treated human HepG2 hepatocellular carcinoma cells 14 and in PC3, a prostate cancer cell line 15. The data suggest that at least two mammalian CAKs are responsible for the phosphorylation of cdk2 and Cdk4/Cdk6 in vivo.
The midblastula transition (MBT), a key developmental event in Xenopus laevis and other animal embryos, was identified by Newport and Kirschner in 1982 16. In zebrafish embryos, MBT occurs at the 512-cell stage, within a narrow 2.5 h window. Since then, zebrafish has become an excellent vertebrate model for studying embryogenesis and for genetic analysis due to their large and transparent embryos and their rapid development, attaining sexual maturity at 3 months of age. In Xenopus and Danio, the MBT is characterized by a loss of cell division synchrony, an increase in cell motility, the onset of wholesale zygotic transcription and the presence of G1 and G2 phases 16, 17; the unique properties of zebrafish make it an ideal model system to study cell cycle and transcription during this special developmental stage.
Over the past decade, much of the research on metazoan Cdk7 and Cyclin H has been conducted in vitro, and in vivo analyses are relatively lacking, especially in the area of embryogenesis. Although the use of dominant-negative mutant has demonstrated that the Cdk7 gene is essential during the midblastula transition in Drosophila embryos 18, the role of Cyclin H has not been fully elucidated. In this study, we explored the role of Cyclin H in zebrafish embryogenesis and development, particularly during MBT. Two novel cDNAs encoding Cyclin H and Cdk7 were cloned from blastula fish embryos. Loss of Cyclin H function by forced expression of a dominant-negative form 19 severely delayed the onset of zygotic transcription in the early embryos and induced apoptosis 5-h post-fertilization (hpf). These results suggest that Cyclin H, similar to Cdk7, facilitates transcription in vivo during MBT 18. Our study also demonstrates that the zebrafish Cyclin H is an essential gene for early larval development.
Materials and Methods
Fish stocks
Zebrafish were raised and maintained according to the standard laboratory conditions 20. Embryos were reared and staged at 28 °C, according to Kimmel et al. 21.
Cloning of zebrafish Cyclin H and Cdk7 cDNA
Cloning of zebrafish full-length Cyclin H and Cdk7 cDNA was performed by 3′ and 5′ smart RACE (Clontech), according to the manufacturer's instructions. Primers used for Cyclin H amplification were the universal amplification primer from the 3′ and 5′ smart RACE kit (UPM 5′-CTA ATA CGA CTC ACT ATA GGG CAA GCA GTG GTA TCA ACG CAG AGT-3′) and nested universal primer (NUP 5′-ACT CTG CGT TGA TAC CAC TGC TT-3′). Primers used for the outer and inner Cyclin H were as follows: for 3′ RACE PCRs, F1, 5′–CGT TCA CAA TCC GTA TCG ACC TTT GGA G-3′ and F2, 5′-CCG AGC AAC AAT GAC AGA TGC CGG ACT G-3′; for 5′ RACE, R1, 5′-CTG TCA TTG TTG CTC GGT TGA GAA AGT-3′ and R2, 5′-CTA AAT CTG TTG TGG GCA CAG CTT GCA T-3′, and R3, 5-CGA TAC GGA TTG TGA ACG ACC AGA TGA A-3′, which were designed based on an EST sequence (Accession Number BM889604). Cloning of Cdk7 was as follows: primers of UPM and NUP were similar to those used for cloning of Cyclin H's, and the primers used for the outer and inner Cdk7 were: for 3′ RACE PCRs, F1, 5′-CGT TCA CAA TCC GTA TCG ACC TTT GGA G-3′ and F2, 5′-CCG AGC AAC AAT GAC AGA TGC CGG ACT G-3′; for 5′ RACE PCRs, R1, 5′-CTG TCA TTG TTG CTC GGT TGA GAA AGT-3′ and R2, 5′-CTA AAT CTG TTG TGG GCA CAG CTT GCA T-3′. Primers were designed based on the EST database sequence (GenBank XM_686667). Amplified fragments were subcloned and sequenced, and the sequences were deposited in the Genbank database under Accession Numbers DQ294346 and DQ294347. RT-PCR was carried out using 5 μg of total RNA isolated from a 6 hpf zebrafish embryo under the following conditions: initial denaturation (95 °C, 3 min), 30 cycles (95 °C, 30 s; 68 °C, 40 s; 72 °C, 2 min) and a final extension (72 °C, 7 min).
Sequence analysis
The amino-acid sequence was determined with DNAMAN program (http://www.lynnon.com/) and ORF Finder (Open Reading Frame Finder) (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Protein sequences were aligned using the CLUSTALX algorithm (ftp://ftp.ebi.ac.uk/pub/software/dos/clustalx/) and Jalview (http://www.jalview.org/Web_Installers/install.htm). Homology searches were carried out using DNAMAN. Construction of the phylogenetic parsimony tree utilized the MEGA software (http://www.megasoftware.net/).
Northern analysis
Cyclin H and Cdk7 PCR fragments were cloned into a pGEM-T-easy vector (Promega). Plasma DNA was linearized (5–10 μg) by digestion with the appropriate restriction enzyme for RNA and DNA probe labeling. DIG RNA Labeling Kit (Roche) was used for labeling of the Cyclin H probe (345 bp), and Cdk7 fragment (370 bp) was labeled with [α-32P]dATP (sp act3000 Ci/mmol, Amersham Pharmacia Biotech) using the Random Primer DNA Labeling Kit (TAKARA). Denaturing formaldehyde agarose gel electrophoresis (10-15 μg of total RNA) was performed as described in Molecular Cloning 22. The RNA was transferred to a positively charged nylon membrane (Hybond N+, Amersham Pharmacia Biotech, UK). After hybridization, Cyclin H was detected by anti-digoxigenin Fab-alkaline phosphatase and stained with BCIP/NBT; for detection of Cdk7 mRNA, the membrane was hybridized with Cdk7 antisense probe, dried and subjected to autoradiography 22.
RT-PCR analysis
Total RNA was isolated from different adult tissues and at different stages of embryogenesis using trizol (Invitrogen). First-strand cDNA was synthesized from 2 μg of total RNA by SuperScript II RT (Life Technologies) according to the manufacturer's protocol. The cDNA was amplified for 30 cycles under the following thermal conditions: 94 °C, 30 s; 52 °C, 30 s; 72 °C, 50 s. Primers for Cyclin H amplification (F362, 5′-CCA GCA CTC AGT TTG TGG-3′ and R853, 5′-TAG CAA ACT CAG CAT AAA TCC-3′) and for Cdk7 (F571, 5′-GGT GTA GGT GTG GAC ATG TGG-3′ and R941, 5′-GTT GAG GAG TTT GGT CTG GGT-3′) were designed according to GenBank Accession Numbers DQ294346 and DQ294347, respectively. Amplification of β-actin was used as an internal control (F, 5′-CCT CCG GTC GTA CCA CTG GTA T-3′ and R, 5′- CAA CGG AAG GTC TCA TTG CCG ATC GTG -3′).
Whole-mount in situ hybridization
Sense and antisense probes of Cyclin H and Cdk7 were generated, as above, by in vitro transcription using SP6 and T7 RNA polymerase, respectively, in the presence of digoxigenin-labeled UTP (DIG RNA Labeling Kit, Roche). Probes of bozozok and no tail were also generated in a similar fashion. Preparation of embryos and hybridization were performed according to Schulte-Merker et al. 23; alkaline-phosphatase-coupled antidigoxigenin Fab-fragments (Roche) were incubated with embryos and larvae and the antibody-antigen complex was detected by staining with BCIP/NBT (Roche). Samples were dehydrated in methanol, cleared and mounted in 2:1 benzyl benzoate: benzyl alcohol.
TUNEL assay
TUNEL assay (TdT-mediated fluorescein-dUTP nick end labeling) was adapted from established protocols 24, using TUNEL Label (Roche) containing fluorescein-dUTP and TdT (terminal deoxynucleotidyl Transferase, Roche). Embryos were washed in TBS (tris-buffered saline) for 5 min at room temperature (RT), fixed in 4% formaldehyde (overnight, 4 °C), washed with TBST (TBS plus 0.1% Tween-20) (3 × 15 min, RT), dechorionated, incubated with TUNEL buffer (Roche, 30 min, RT), and then incubated in 50 μl TUNEL reaction mixture (1 h, 37 °C, in the dark). The reaction was stopped by washing samples with TBST (5 × 5 min, RT); the samples were blocked in TBST containing 1% BSA (1 h, RT), incubated with anti-fluorescein antibody coupled to alkaline phosphate (TUNEL AP, Roche, overnight, 4 °C), mounted with TBST/glycerol (1:1) and detected by BCIP/NBT.
Capped RNA transcription and injection experiments
The truncated Cyclin H (CTΔ257) was selected for dominant-negative Cyclin H experiments according to deletion mutant analyses described by Andersen et al. 19. The coding sequences for the dominant-negative form of Cyclin H or the GFP ORF were amplified using primers that introduce EcoRI and XhoI restriction sites. The resulting fragments were subcloned into pCS2+ and transcribed using the Ambion Message Machine kit. Typically, 0.3–0.5 ng of dominant-negative Cyclin H mRNA was injected into the yolk cell of one-cell stage embryos. Where appropriate, 0.1 ng of GFP mRNA was injected as a lineage tracer and 0.3 ng of GFP mRNA as the negative control. Phenol red (0.05%) was co-injected as a non-toxic injector tracer.
Results
cDNA cloning and sequence analysis of zebrafish Cyclin H and Cdk7 genes
Primers were designed according to the zebrafish EST sequence, given its high 3′ similarity to human Cyclin H (Accession No. BM889604). Following RT-PCR using RNAs isolated from shield stage embryos (6 hpf), the full-length Cyclin H cDNA was cloned by 3′ and 5′ smart RACE and nested PCR using cDNAs obtained from 6 hpf embryos. Sequencing analysis of the RACE fragment revealed a Cyclin H consensus sequence of 1 302 bp containing an ORF of 957 bp, which was submitted to the GenBank (Accession No. DQ294346). The full-length cDNA for Cdk7 was cloned using a similar strategy, revealing a consensus sequence of 1260 bp with an ORF of 1035 bp (Accession No. DQ294347). The Cyclin H ORF encodes a deduced polypeptide of 319 amino acids, confirming that the cDNA indeed corresponds to zebrafish Cyclin H 19, although the protein only shares 65% identity with the human ortholog (Figure 1A). We were unable to find other isoforms by 3′ and 5′ smart RACE using RNAs from 6 hpf embryos or ovary, suggesting that only one major form of Cyclin H message is present in zebrafish. The zebrafish Cdk7 ORF encodes a 345 amino-acid polypeptide with 86% identity to the human ortholog (Figure 1B). The sequence alignment of Cyclin H and Cdk7 demonstrates that they are conserved in vertebrates from fish to mammals (Figure 2A and 2B). Phylogenetic parsimony tree analysis (Figure 2A and 2B) also suggests that Cyclin H might have evolved at a more rapid pace than that noted for Cdk7, which may be associated to their respective functions. We searched the database (http://pre.ensembl.org/Danio_rerio/blastview) using the zebrafish cyclin H and cdk7 cDNAs separately, and found that both genes are located at chromosome 5 and represent single copy genes.
Alignment of zebrafish Cyclin H and Cdk7 with human Cyclin H (A) and Cdk7 (B). The amino-acid sequences of human Cyclin H (NM_001239) and Cdk7 (NM_001799) were aligned with the zebrafish Cyclin H and Cdk7 sequences, respectively. Numbers at the right side refer to the amino-acid position. Asterisks indicate conserved residues in all sequences; a single dot indicates semi-conserved substitutions and a double dot indicates conserved substitutions.
Comparison of amino-acid alignment of Cyclin H sequences among zebrafish, Homo sapiens (NM_001239), X. laevis (BC106342), Gallus gallus (XP_424908), Drosophila (NM_079483), C. elegans (P90866) and S. cerevisiae (P39073) (A). Zebrafish Cdk7 and Cdk7 sequences of Homo sapiens (NM_001799), X. laevis (NM_001017219), Gallus gallus (XP_424761), Drosophila (DMU56661), C. elegans (AAD38186) and S. cerevisiae (CAA98675) (B) were used to construct a phylogenetic parsimony tree with MEGA software. Statistical reliability of the nodes was obtained by bootstrap analysis (1 000 replications) with the parsimony maximum method.
Northern analysis of Cyclin H and Cdk7
Given that only a single copy of the Cyclin H or Cdk7 gene was detected, RNA samples from shield stage embryos and adult ovary (data not shown) were chosen for Northern blot analysis. Consistent with full-length cDNA cloning results, in each case a major transcript with the predicted size was identified (Figure 3A and 3B).
Northern blot analysis of total RNAs from the shield stage embryos using a digoxigenin-labeled Cyclin H probe (A) and a [32P]-labeled Cdk7 probe (B). Transcripts size (arrow) of Cyclin H and Cdk7 was consistent with that of the cloned full-length cDNAs. RNA denaturing formaldehyde agarose gel was used to quantitate 28 s and 18 s rRNAs. Total RNAs were loaded onto 1.2% agarose gel.
RT-PCR analysis of Cyclin H and Cdk7 expression patterns
RT-PCR analysis was performed to investigate zebrafish Cyclin H and Cdk7 mRNA expression during embryogenesis. Expression of Cyclin H was identified in the ovary (Figures 4 and 5), raising the possibility that Cyclin H mRNA might be maternally inherited. This was later confirmed by the detection of its message in unfertilized eggs (Figure 6A). Additionally, bands corresponding to the predicted size of Cyclin H fragment were detected during all examined stages of embryo development, from 2 hpf to 5 dpf (Figure 4), and the expression pattern of Cdk7 was found to be similar to that of Cyclin H (data not shown). We further investigated the expression of Cyclin H and Cdk7 in various adult tissues including the brain, fin, ovary, testis, skin, gill, intestine, muscle, kidney, heart and liver (Figure 5). The expression of Cyclin H was generally higher than that of Cdk7 in all chosen tissues, although a strict quantitative analysis was not performed. Additionally, samples from the ovary, testis and intestine consistently appeared to have the highest levels of Cyclin H and Cdk7 mRNAs, while the brain, fin, eye and kidney had relatively lower levels, with little Cdk7 mRNA identified in the kidney.
Expression of zebrafish Cyclin H during embryonic development assayed by RT-PCR using total RNAs prepared from the indicated stages. Amplification of β-actin was used as an internal control. The estimated size of PCR products for β-actin and Cyclin H was 350 and 491 bp, respectively (n = 3 parallel experiments). h, hours post fertilization; d, days post fertilization.
Expression of zebrafish Cyclin H and Cdk7 in various adult tissues. RT-PCR analysis was performed using RNAs isolated from the brain (b), fin (f), eye (e), ovary (o), testis (t), skin (s), gill (g), intestine (i), muscle (m), kidney (k), heart (h) and liver (l). β-Actin was used as an internal control. The estimated size of PCR products for β-actin, Cyclin H and Cdk7 was 350, 491 and 371 bp respectively. DNA marker was DL2000.
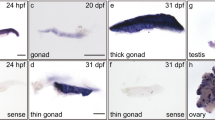
Whole-mount ISH analysis of zebrafish Cyclin H (A-O) and Cdk7 (P) transcripts during different embryonic developmental stages. mRNA of Cyclin H was identified in unfertilized eggs (A). Control hybridization with a sense probe of Cyclin H at the 2-cell stage is shown in (D). At early developmental stages, Cyclin H expression was ubiquitous and uniform (A–M). Note the obvious moving of Cyclin H (arrow in C, E). Shown are lateral view (A-E, H-J), dorsal view (F, G, N), lateral view, anterior to left (M, O, P) and lateral view, anterior to right (K, L). Strong Cyclin H staining is noted in the anterior neural tube, brain and eye at 24 hpf. Gradually increased and prominent expression of Cyclin H and expression of Cdk7 is dominant in the anterior neural tube, brain, eye, liver and heart by 5 dpf.
Spatial expression pattern of Cyclin H and Cdk7 during embryogenesis analyzed by in situ hybridization
Sense and antisense probes of Cyclin H and Cdk7 were used to detect the spatial expression patterns of Cyclin H and Cdk7 during embryogenesis, by in situ hybridization (ISH). While no signal was noted in control hybridizations with sense probes (Figure 6D), mRNA of Cyclin H was identified in unfertilized eggs (Figure 6A), 2-cell (Figure 6B and 6C), 4-cell (Figure 6-F), 8-cell (Figure 6G and H), 2 hpf (Figure 6I), 6 hpf (Figure 6J), 10 hpf (Figure 6K), 15 hpf (Figure 6L), 24 hpf (Figure 6M) and 5 dpf embryos (Figure 6N and 6O). While ubiquitous Cyclin H mRNA distribution was noted in the whole embryos before 24 hpf (Figure 6A-L), at 24 hpf, the expression became more restricted to the brain rudiment (Figure 6M). In 5 dpf embryos, the expression was less diffuse (Figure 6N and 6O), becoming restricted in the developing brain, eye, liver and heart (Figure 6N and 6O). Although Cyclin H expression levels were much higher in the ovary and testis of adult zebrafish (Figure 5), its expression level during gonadogenesis was unclear 25. The pattern of Cdk7 expression overlapped with that of Cyclin H at 5 dpf, but it was more restricted, particularly in the developing brain, eye, liver and heart (Figure 6P).
Effect of dominant-negative Cyclin H on early embryo development
A dominant-negative form of Cyclin H was used to study the function of the zebrafish Cyclin H gene, as it has been demonstrated to specifically compete with endogenous Cyclin H for Cdk7 binding while losing its regulatory function 19. Injection of Cyclin HDN mRNA (0.5 ng) generated developing embryos morphologically arrested (36% fating to death) at the midblastula stage (Figure 7B and Table 1). Apoptosis within these embryos, as detected by the TUNEL assay, was noted at 5 hpf (data not shown), and continued at 6 hpf (Figure 7F) and 8 hpf (data not shown), with embryo death seen at the 90% epiboly stage (Table 1). No effects were seen in GFP-injected controls. About 48% of Cyclin HDN-injected embryos showed delayed development (data not shown), and they continued to develop (fating to being abnormal), albeit the development was slower than GFP-injected controls. Comparison of developmental rates suggested that while control embryos developed to the 90%-epiboly stage, these Cyclin HDN-injected embryos had only attained the shield stage (Figure 7C and 7D). For these 48% of Cyclin HDN-injected embryos that continued the development through 60 hpf (Table 1, Figure 8B), abnormal embryo configuration, with severe defects in head and eye development as well as small size body, was noted, compared to GFP-injected controls at 30 hpf (Figure 8A and 8B). Developmental aberrations in these embryos were noted particularly in the eye, with defects such as the development of only one eye (Figure 8B) or no eye (Figure 8C) or fusion of both eyes (Figure 8D). No such effects on embryonic development were seen with control injection of the GFP mRNA, as the control embryos continued through gastrula and bud stages, similar to uninjected embryos (Table 1).
Expression of a dominant-negative form of Cyclin H resulted in embryo arrest and delay during development and induced apoptosis at the early gastrulation stage. mRNAs encoding Cyclin H DN (B, D, F) and the GFP control (A, C, E) were injected into embryos at 1- or 2-cell stage, and the embryos were allowed to develop to the shield and bud stages. Samples were harvested at the shield stage for TUNEL Assay. Apoptotic bodies are indicated (black arrows). All embryos are shown in lateral views with dorsal to the right.
Cyclin H loss-of-function during Zebrafish development. RNAs encoding Cyclin HDN with GFP and GFP control were injected into embryos at the 1- or 2-cell stage; the embryos were then grown to 30 hpf (A) and 60 hpf (B, C, D) and analyzed for the presence of morphological defects. Abnormal body shape and eyes and brain were observed in Cyclin HDN-injected embryos.
To investigate whether the defects noted above were indeed caused by Cyclin HDN, and whether Cyclin HDN disturbed the transcription, we analyzed bozozok (boz) and no tail (ntl) genes, which initiate their expression between the MBT and the onset of gastrulation. Cyclin HDN-treated embryos showed ectopic and reduced bozozok and no tail expression throughout the blastoderm, respectively; embryos shown were at sphere stage (4 hpf) (Figure 9A and 9B) and shield stage (6 hpf) (Figure 9C and 9D). Expression of boz and ntl was initiated in Cyclin HDN-injected embryos, but their normal restricted expression patterns were disrupted compared to GFP-injected controls. The expression of boz was scattered and no longer restricted to the dorsal region (Figure 9B); the expression of ntl was found in scattered positive cells around the margin and central clearing (Figure 9D).
Expression of Cyclin HDN disturbed the onset of zygotic transcription. Expression of bozozok (A, B) and no tail (C, D) were detected by whole-mount ISH in GFP-infected control and Cyclin HDN-injected embryos. All embryos are shown with the animal pole facing the viewer at the onset of the sphere stage (4 hpf) (A, B) and shied stage (6 hpf) (C, D); dorsal is to the right.
Discusssion
Recent research has identified two non-overlapping sets of Cdks in metazoans: Cdk1, Cdk2, Cdk4 and Cdk6, dedicated to cell-division control, and Cdk8 and Cdk9, primarily implicated in transcription 26. However, the Cdk7 complex cannot be easily classified 26 as Cdk7/Cyclin H has a dual role: first, as the CAK of cell cycle Cdks, and second, as a component of the general transcription factor TFIIH, phosphorylating the CTD of RNA polymerase II.
Recent research has challenged the generally accepted idea that cell-division Cdks, such as Cdk1, Cdk2, Cdk4 and Cdk6, and their different cyclin partners, are essential for cell function. Studies have demonstrated normal fetal development following disruption of the D- and E-type cyclins, Cdk4, Cdk6 and Cdk2, suggesting that the individual functions of these genes were not essential for cell cycle progression 27, 28. One possible explanation is that cyclins that promote entry into the S phase in yeast are deleted without major consequences because mitotic cyclins can take over their function. This cyclin complementation has been demonstrated in mammalian cells 29. Additionally, much of what we know regarding the cell cycle has been derived from studies on single-celled organisms, such as yeast, and on tissue culture cells, systems that do not necessarily reflect the inherent complexity of regulation required by a multicellular organism like the early embryo 30. This is why in this paper we chose to study Cyclin H and Cdk7 function in vivo during zebrafish embryo development.
We isolated cDNAs coding for the zebrafish Cyclin H and Cdk7, investigated the effect of Cyclin H on early embryo development and characterized its role in MBT. The deduced zebrafish Cyclin H ORF encodes a polypeptide of 319 amino acids, suggesting that the conserved motifs in zebrafish Cyclin H are similar to those found in mammals 19 (Figure 1A). We were unable to identify any additional isoforms, suggesting that only one major form of Cyclin H is present in zebrafish. The zebrafish Cdk7 ORF encodes a 345 amino-acid polypeptide with 86% identity to the human ortholog and a highly conserved T170 18 (Figure 1B). We searched the database (http://pre.ensembl.org/Danio_rerio/blastview) using the zebrafish cyclin H and cdk7 cDNAs separately and found that both genes are located at chromosome 5 and represent single copy genes. Northern blot analysis of Cyclin H and Cdk7 indicated the presence of a major transcript with the predicted size for each gene in 6 hpf embryos (Figure 3A and 3B), consistent with our cloning results. We generated antibodies against Cyclin H and Cdk7 and confirmed the deduced molecular weight of each protein by Western blot analysis (data not shown).
To assess whether Cyclin H and Cdk7 mRNAs were present during zebrafish embryonic development, RT-PCR (Figures 4 and 5) and ISH (Figure 6) were performed with embryos at different developmental stages and with various tissue types. During early embryogenesis, Cyclin H mRNA was homogeneously distributed, which may have been the result of maternal inheritance (Figure 6A). Expression of Cyclin H in the ovary also suggested that Cyclin H mRNA was maternally inherited. The expression pattern of Cdk7 during embryogenesis was similar to that of Cyclin H (data not shown). At 24 hpf, Cyclin H expression gradually became more restricted to the brain rudiment (Figure 6M) with greater restriction noted in the developing brain, eye, liver and heart from 24 hpf to 5 dpf (Figure 6N and 6O). The pattern of Cdk7 expression was more restricted than Cyclin H at 5 dpf, shown mainly in the developing brain, eye, liver and heart (Figure 6P). This progressively restricted pattern of expression was similar to that reported for X. laevis, in which Cdk1, Cdk2, Cdk4 and cyclins D1, D2, E, A1, A2 and B1 mRNA expression became tissue specific after gastrulation, as they exhibited distinct expression patterns and were not highly expressed in all regions of active proliferation 31.
The similarity in expression patterns between the different cyclins and cdks suggests that, in early embryos, cyclins and cdks are required at high levels in all cells, and proliferation is fairly uniform prior to gastrulation; however, soon after gastrulation, stereotypical, region-specific patterns were observed 32. Although Cyclin H expression during gonadogenesis (5 dpf or later) is not fully understood 25, RT-PCR data suggest that its expression is much higher in the ovary and testis of adult zebrafish, consistent with the finding that meiotic expression of Cyclin H/Cdk7 is responsible for activating cyclin A1/Cdk2 in male germ cells in mouse 33. In addition, transcription of Cyclin H in rat neurons was increased primarily after ischemia, suggesting additional roles for Cyclin H in neurons, other than cell cycle regulation and DNA repair 34.
Injection of Cyclin HDN mRNA into embryos resulted in two distinct phenotypes. First, about 36% of these embryos displayed morphological arrest at a midblastula stage (Figure 7A and 7B), with apoptosis first observed at 5 hpf (data not shown) and with mutant death at the tail bud stage (Table 1), compared to GFP-injected controls, which developed normally. Typically, apoptosis in zebrafish embryos first appears at 10 hpf 35, suggesting that injection of Cyclin HDN disturbed normal cell cycle or transcription, inducing apoptosis through activation of the maternally inherited apoptosis machinery 36. Our data suggest that apoptosis may have been the cause of death of these embryos at the 8-10 hpf stage. The second phenotype observed in about 48% of embryos was the delay of early embryogenesis, although these delayed embryos eventually reached the midblastula stage and continued their development to 60 hpf (Table 1). Given that cyclin A also interacts with and activates Cdk7 19, cyclin A compensation may have occurred after MBT. Although these embryos reached 60 hpf, they were abnormal, with severe defects in head and eye development and with small size bodies compared to GFP-injected controls (Figure 8A-D and Table 1). These defects may have resulted from cell death in tissues where higher Cyclin H expression levels were observed. To investigate if the above phenotype was caused by the disturbance of initiation of zygotic transcription owing to the fact that Cdk7/Cyclin H functions as components of the general transcription factor TFIIH 26, we tested the expression of boz and ntl in Cyclin HDN mRNA-injected embryos after allowing them to develop to the sphere stage and shield stage, respectively. We found that the expression of boz and ntl was reduced and their normal restricted expression pattern was disrupted (Figure 9A-D). These data suggest that Cyclin HDN protein disturbed early zygotic transcription at MBT, similar to that observed in Cdk7 mutants 18. Thus, the injected embryos display a defect of uncoordinated initiation of zygotic gene expression after the MBT.
This is, to our knowledge, the first study of cloned Cyclin H and Cdk7 in Danio rerio. Our results suggest that zebrafish Cyclin H is functional and plays a key role during early fish development.
References
Shuttleworth J . The regulation and functions of Cdk7. Prog Cell Cycle Res 1995; 1:229–240.
Lolli G, Johnson LN . CAK-Cyclin-dependent activating kinase: a key kinase in cell cycle control and a target for drugs? Cell Cycle 2005; 4:572–577.
Garrett S, Barton WA, Knights R, et al. Reciprocal activation by cyclin-dependent kinases 2 and 7 is directed by substrate specificity determinants outside the T loop. Mol Cell Biol 2001; 21:88–99.
Ossipow V, Tassan JP, Nigg EA, Schibler U . A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell 1995; 83:137–146.
Martinez AM, Afshar M, Martin F, et al. Dual phosphorylation of the T-loop in Cdk7: its role in controlling Cyclin H binding and CAK activity. EMBO J 1997; 16:343–354.
Fisher RP, Jin P, Chamberlin HM, Morgan DO . Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell 1995; 83:47–57.
Busso D, Keriel A, Sandrock B, et al. Distinct regions of MAT1 regulate Cdk7 kinase and TFIIH transcription activities. J Biol Chem 2000; 275:22815–22823.
Inamoto S, Segil N, Pan ZQ, Kimura M, Roeder RG . The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, targets and enhances CAK activity on the POU domains of octamer transcription factors. J Biol Chem 1997; 272:29852–29858.
Cismowski MJ, Laff GM, Solomon MJ, Reed SI . KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol 1995; 15:2983–2992.
Espinoza FH, Farrell A, Erdjument-Bromage H, Tempst P, Morgan DO . A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science 1996; 273:1714–1717.
Lee KM, Saiz JE, Barton WA, Fisher RP . Cdc2 activation in fission yeast depends on Mcs6 and Csk1, two partially redundant Cdk-activating kinases (CAKs). Curr Biol 1999; 9:441–444.
Shimotohno A, Matsubayashi S, Yamaguchi M, Uchimiya H, Umeda M . Differential phosphorylation activities of Cdk-activating kinases in Arabidopsis thaliana. FEBS Lett 2003; 534:69–74.
Kaldis P, Solomon MJ . Analysis of CAK activities from human cells. Eur J Biochem 2000; 267:4213–4221.
Nagahara H, Ezhevsky SA, Vocero-Akbani AM, et al. Transforming growth factor beta targeted inactivation of cyclin E:cyclin-dependent kinase 2 (Cdk2) complexes by inhibition of Cdk2 activating kinase activity. Proc Natl Acad Sci USA 1999; 96:14961–14966.
Ukomadu C, Dutta A . Inhibition of cdk2 activating phosphorylation by mevastatin. J Biol Chem 2003; 278:4840–4846.
Newport J, Kirschner M . A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell 1982; 30:675–686.
Kane DA, Kimmel CB . The zebrafish midblastula transition. Development 1993; 119:447–456.
Leclerc V, Raisin S, Leopold P . Dominant-negative mutants reveal a role for the Cdk7 kinase at the mid-blastula transition in Drosophila embryos. EMBO J 2000; 19:1567–1575.
Andersen G, Busso D, Poterszman A, et al. The structure of Cyclin H: common mode of kinase activation and specific features. EMBO J 1997; 16:958–967.
Monte Westfield. The Zebrafish Book. Eugene: University of Oregon Press, 1995.
Kimmel CB, Ballard WW, Kimmel SR, et al. Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203:253–310.
Joseph Sambrook DR . Molecular Cloning: A Laboratory Manual. Dallas: Cold Spring Harbor Laboratory Press, 2001.
Schulte-Merker S, Ho RK, Herrmann BG, Nusslein-Volhard C . The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 1992; 116:1021–1032.
Chan DW, Yager TD . Preparation and imaging of nuclear spreads from cells of the zebrafish embryo. Evidence for large degradation intermediates in apoptosis. Chromosoma 1998; 107:39–60.
Solnica-Krezel L . Pattern Formation in Zebra Fish. Berlin: Springer Publishing Company, 2002.
Fisher RP . Secrets of a double agent: Cdk7 in cell-cycle control and transcription. J Cell Sci 2005; 118:5171–5180.
Geng Y, Yu Q, Sicinska E, et al. Cyclin E ablation in the mouse. Cell 2003; 114:431–443.
Sherr CJ, Roberts JM . Living with or without cyclins and cyclin-dependent kinases. Genes Dev 2004; 18:2699–2711.
Mendez J . Cell proliferation without cyclin E-Cdk2. Cell 2003; 114:398–399.
Richard-Parpaillon L, Cosgrove RA, Devine C, Vernon AE, Philpott A . G1/S phase cyclin-dependent kinase overexpression perturbs early development and delays tissue-specific differentiation in Xenopus. Development 2004; 131:2577–2586.
Vernon AE, Philpott A . The developmental expression of cell cycle regulators in Xenopus laevis. Gene Expr Patterns 2003; 3:179–192.
Saka Y, Smith JC . Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol 2001; 229:307–318.
Kim JM, McGaughy JT, Bogle RK, Ravnik SE . Meiotic expression of the Cyclin H/Cdk7 complex in male germ cells of the mouse. Biol Reprod 2001; 64:1400–1408.
Jin K, Nagayama T, Chen J, et al. Molecular cloning of a cell cycle regulation gene Cyclin H from ischemic rat brain: expression in neurons after global cerebral ischemia. J Neurochem 1999; 73:1598–1608.
Yabu T, Kishi S, Okazaki T, Yamashita M . Characterization of zebrafish caspase-3 and induction of apoptosis through ceramide generation in fish fathead minnow tailbud cells and zebrafish embryo. Biochem J 2001; 360:39–47.
Ikegami R, Hunter P, Yager TD . Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev Biol 1999; 209:409–433.
Acknowledgements
We thank Yihong Wang for expert fish care and Shifeng Li for Northern of Cdk7. This study was supported by a grant from E-Institutes of Shanghai Municipal Education Commission, Project Number E03003 and a grant from Shanghai Municipal Commission for Science and Technology (Grant No. 04DZ14006).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, Q., Wu, Z., Lv, W. et al. Developmental expression of Cyclin H and Cdk7 in zebrafish: the essential role of Cyclin H during early embryo development. Cell Res 17, 163–173 (2007). https://doi.org/10.1038/sj.cr.7310144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7310144
Keywords
This article is cited by
-
GATA4 as a novel regulator involved in the development of the neural crest and craniofacial skeleton via Barx1
Cell Death & Differentiation (2018)
-
Genetic and pharmacological inhibition of CDK9 drives neutrophil apoptosis to resolve inflammation in zebrafish in vivo
Scientific Reports (2016)
-
Low Expression of CyclinH and Cyclin-Dependent Kinase 7 Can Decrease the Proliferation of Human Esophageal Squamous Cell Carcinoma
Digestive Diseases and Sciences (2013)
-
Molecular cloning and expression analysis of xpd from zebrafish (Danio rerio)
Molecular Biology Reports (2012)
-
Global proteomics analysis of testis and ovary in adult zebrafish (Danio rerio)
Fish Physiology and Biochemistry (2011)