ABSTRACT
Incubation of dinoflagellate Crythecodinium cohnii chromosomes in cytoplasmic extracts of unfertilized Xenopus laevis eggs resulted in chromosomes decondensation and recondensation, nuclear envelope assembly, and nuclear reconstitution. Dinoflagellate Crythecodinium cohnii is a kind of primitive eukaryote which possesses numerous permanently condensed chromosomes and discontinuous double-layered nuclear membrane throughout the cell cycle. The assembled nuclei, being surrounded by a continuous double membrane containing nuclear pores and the uniformly dispersed chromatin fibers are morphologically distinguishable from that of Dinoflagellate Crythecodinium cohnii. However, incubation of dinoflagellate Crythecodinium cohnii chromosomes in the extracts from dinoflagellate Crythecodinium cohnii cells does not induce nuclear reconstitution.
Similar content being viewed by others
IINTRODUCTION
Dinoflagellates are a group of diverse eukaryotic protists, possessing a number of peculiar traits of nuclear organization, suggestive for the transition between the prokaryotic and eukaryotic state. Among these are: (a) permanently condensed chromosomes during the whole cell cycle1, (b) chromatin devoid of histone and nucleosomes2, (c) very complex cell cycle3. These prokaryotic affinities of the dinoflagellates could indicate their closeness to the ancestral state of all present-day eukaryotes, at least regarding nuclear organization4. Molecular data suggest that Crythecodinium cohnii is one of the most primitive of the dinoflagellate Peridiniales5. How can dinoflagellate chromosome keep highly condensed when replication, transcription and translation are proceeding during the whole cell cycle? What are the important components, DNA, histone or nonhistone in C cohnii? However, very few studies about nuclear organization of dinoflagellate have been done.
Eggs of Xenopus laevis have proved to be exceptionally favorable sources of cell-free systems for studying the assembly of the cell nucleus. Xenopus laevis eggs store abundant protein and membrane components required to generate a functional nuclear envelope around exogenously added DNA or chromatin6,7. Egg extracts can be stably prepared in either interphase or metaphase. Interphase extracts can support nuclear assembly and transport, whereas mitotic extracts can disassemble nuclei. Since 1989, in our laboratory, we have successfully got the nuclei assembled in vitro around exogneous naked DNA from animals8, plants9, rDNA10 and demembranated sperm chromatin11. Meanwhile, we have done some research concerning the mechanism of nuclear envelope reconstitution12. We wonder whether the intact dinoflagellate chromosomes can be reconstituted or not in a cell-free system derived from Xenopus laevis. Here we reported the details of Crythecodinium cohnii chromosomes being reconstituted in a cell-free system of Xenopus laevis eggs, followed by nuclear reassembly. Differences of the nuclei and nuclear envelope between that of dinoflagellate and assembled nuclei were also reported.
MATERIALS AND METHODS
Cell culture
Crythecodinium cohnii was kindly provided by Dr. Joseph T.Y. Wong (Hong Kong University of Science and Technology). Strains were maintained on 1.5 % MLH agar medium according to the method of Tuttle and Loeblech13. For intensive cultures, clones were subcultured in MLH liquid medium and put in the dark at 27°C.
Isolation of Dinoflagellate C. cohnii chromosomes
Dinoflagellate C. cohnii cells were harvested by centrifugation for 5 min at 3,000 g and suspended in isolation medium (0.75 M sucrose, 2.5 % w/v Ficoll, 5 mM CaCl2, 0.5 mM DTT, 0.01 M Tris-HCl, pH7.5, and 0.1 mM PMSF). Cells were disrupted briefly in a W-220 ultrasonicator. Nuclei, the brown pellet, sendimented from the homogenate by centrifugation at 2,000 g for 5 min, and homogenized in isolation medium containing 0.05 % (v/v) Triton X-100 using a Potter-Elvehjem homogenizer. The homogenate was layered over a 2-step discontinuous gradient consisting of the following: 2.4 M sucrose, 5 mM CaCl2 and 0.01 M Tris-HCl (pH 7.5), overlayered with 2.2 M sucrose, 5 m M CaCl2 and 0.0l M Tris-HCl (pH 7.5). The homogenate were centrifuged at 48,000 g for 1 h. The pellet was suspended with isolation buffer containing lysolecithin at 330mg/μl for 10 min, and centrifuged at 3,000 g for 10 min. The pellet was chromosome extracts. The extracts were washed with ice-cold isolation buffer containing 3 % bovine serum albumin (BSA), and then with isolation buffer containing 0.4 % BSA for three times, finally resuspended in isolation buffer with 30 % glycerol and stored at −70°C.
Preparation of Xenopus egg extracts
Healthy Xenopus laevis females were injected with 600 units of human chorionic gonado-tropin (Sigma) the night before use and were placed in 5 liters of 100 mM NaCl. Eggs were dejellied with 2 % cysteine (pH 7.8) , rinsed twice in MMR (0.1 M NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 5 mM Hepes, 0.1 mM EDTA), and activated by incubation in a solution containing MMR, 5 % Ficoll, and 0.2 μg/ml of the Ca2+ ionophore A23187. The eggs were rinsed three times with 1/4 (MMR plus 5 % Ficoll and were allowed to incubate another 20 min. Following this the eggs were rinsed three times with three volumes of a solution containing 250 mM sucrose, 2.5 mM MgCl2, 50 mM KCl, 100 mg/ml cycloheximide, 5 μg/ml cytochalasin B, and 1 mM dithiothreitol. Then the eggs were crushed at 10,000 g, 4°C for 15 min. The material between the top lipid layer and yolk pellet was removed and centrifuged again at 10,000 g, 4°C for 15 min. The supernatant was used as egg extracts.
Preparation of dinoflagellate C. cohnii cell extracts
Dinoflagellate C. cohnii cells were harvested by centrifugation for 5 min at 3,000 g and suspended in 1/2 volume of a solution containing 250 mM sucrose, 2.5 mM MgCl2, 50 m M KCl, 100 mg/ml cycloheximide, 5 μg/ml cytochalasin B, and 1 mM dithiothreitol. The cells were crushed in a W-220 ultrasonicator. The homogenate was centrifuged at 10,000 g, 4°C for 15 min. The supernatant was centrifuged at 100,000 g, 4°C for 1 h. The supernatant was used as dinoflagellate C. cohnii cell extracts.
Nuclear assembly in vitro
8 μl dinoflagellate C. cohnii chromosomes extracts were mixed with 100 μl freshly prepared X. Laevis egg extracts or 100 μl dinoflagellate C. cohnii cell extracts respectively. Then the ATP regenerating system (2 mM ATP, 50 μg/ml creatine kinase, 2 mM phosphocreatine) was added. The mixture was incubated at 22°C.
Fluorescent microscopy
After incubation, the samples were fixed in 2.5 % glutaraldehyde, stained with DAPI (DNA dye), and examined by florescent microscopy.
Electron microscopy
Aliquots of the incubation mixture were fixed in ice-cold 2 % glutaraldehyde and osmium tetroxide successively, dehydrated through a graded concentration series of acetone and then embedded in Epon 812. Ultrathin sections were cut, stained with uranyl acetate and lead citrate, and finally examined under JEM-100CX transmission electron microscope.
RESULTS
Successful isolation of the dinoflagellate C. cohnii chromosomes has been scarcely reported because dinoflagellate C. cohnii possesses a thick theca and numerous “starch” granules which make it very difficult to isolate its chromosomes. Thus, we first isolate the nuclei as described by Rizzo14. Then, lysolecithin was used to remove nuclear envelope effectively and to get the chromosome extracts.
The unique structure of the chromosomes of dinoflagellate Crypthercodinium cohnii
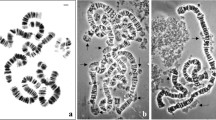
Dinoflagellate C. cohnii possesses 99-100 rod-like acentric chromosomes per cell, which keep condensed during the whole cell cycle (Fig 1). When they were incubated with Xenopus laevis egg extracts, some interesting changes were happened: chromosomes decondensation, recondensation, followed by nuclear reconstitution in steps. The highly condensed dinoflagellate C. cohnii chromosomes could be stained with DAPI (Fig 2A). At the ultrastructral level, there were numerous arched whirls of chromosomal fibrils in a single C. cohnii chromosome (Fig 3A), which is very different from those of higher eukaryotic metaphase chromosomes, but similar to those of bacterial nucleoids. The diameter of dinoflagellate chromosomes is about 30-50 nm.
The ultrastructure of dinoflagellate C. cohnii.
(A) Dinoflagellate C. cohnii cells were fixed, embedded, and sectioned for electron microscopic observation. N: nucleus. Bar=5 μm
(B) Inset: the nucleus of dinoflagellate C. cohnii was stained with DAPI (DNA dye). Note the dot-like permanently condensed chromosomes. Bar=2 μm
Stages of Nuclear reconstitution of C. cohnii chromosomes in X. Laevis egg extracts A mixture containing 100 μl of Xenopus egg extracts, 2 m M ATP, 20 m M creatine phosphate, 50μg/ml creatine kinase, and 8 μl of Dinoflagellate C. cohnii DNA was incubated at 22°C. At the end of incubation (A) 0 min, (B) 20 min, (C) 60 min, (D) 90 min, (E) 150 min, and (F) 180 min, aliquots of the mixture were removed, stained with the DNA-specific flourescent dye DAPI, and observed with a flourescence microscope at a magnification of 1,000 × During the 180 min incubation, the initial Dinoflagellate C. cohnii chromosomes (A) became elongated and swelled (B), then decondensed (C), recondensed (D), reorganized as a packed sphere (E), and (F) assembled into a nucleus. Bar=1 μm
Electron micrograph of Dinoflagellate C. cohnii chromosomes at different stages of nuclear reconstitution. Nuclei were reconstituted around Dinoflagellate C. cohnii chromosome for 180 min, as described in Material and Methods, and then fixed, embedded, and sectioned for electron microscopic observation. (A) At the beginning of incubation, Dinoflagellate C. cohnii chromosomes keep highly condensed. One chromosome was showed. Note the peculiar arrangement of chromosomal fibers. (B) During the first 20 min of incubation, chromosomes were elongated and swelled. (C) At 60 min, rod-like elongated chromosomes were decondensed to assume chromatin-like structures. There were many membrane vesicles (arrow indicated) around it. (D) Following the recondensation of chromatin-like structures, the vesicles fused into double-layered membrane fragments which can be seen in the upper part of the figure. (E) The chromatin-like structure was reorganized into packed spheres.
(F) At 180 min, the packed spheres were encapsulated within a well-defined membrane.
(A) and (B): Bar=0.2mm (C), (D), (E) and (F): Bar=2 μm
Nuclear assembly of C. cohnii chromosomes in cell-free system from Xenopus laevis eggs
Stage I. This stage covered in the first 20 min of incubation. The chromosomes began to elongate and swell, showing discernible decondensation but still keeping rod-like in shape (Fig 2B, 3B).
Stage II. This stage covered the period between 20-60 min of incubation. The chromosomes decondensed continuously so that they no longer maintained their rod-like shape, but displayed some chromatin-like fibrils (Fig 2C, 3C). Meanwhile, there were many membrane vesicles appeared around the chromatin-like structures (Fig 3C, arrow).
Stage III. This stage covered the period between 60-90 min of incubation. The chromatin-like structures became recondensed, and many of them reorganized into tightly packed spheres (Fig 2D). This was followed by the gradual fusion of the membrane vesicles into double-layered membrane segments around the chromatin-like structure and packed spheres (Fig 3D).
Stage IV. This stage covered the period between 90-150 min of incubation. The chromatin-like structures condensed continuously until they were contained within a membrane structure (not fully complete) which was larger in volume than the initial highly packed sphere (Fig 2E, 3E compared with Fig 2D, 3D).
Stage V. This stage covered the period between 150-180 min, of incubation The packed spheres acquired a continuous nuclear envelope with pores and became assembled nuclei (Fig 2F, 3F). The chromatin in assembled nuclei was dispersed uniformly at this stage.
The newly assembled nuclei were morphologically different from those of in dinoflagellate cells (compare Fig 1B with Fig 2F). Electron micrograph showed that the nuclear envelope of dinoflagellate C. cohnii is discontinuous (Fig 4A). However, the reconstituted nuclei possess a well-defined membrane with nuclear pores (Fig 4B) which is morphologically indistinguishable from that of normal higher eukaryotes interphase nuclei. Contrary to the highly condensed chromosomes being attached to the dinoflagellate C. cohnii nuclear envelope, the chromatin in the newly assembled nuclei disperses uniformly, similar to that of typical higher eucaryotic interphase nuclei, in which chromatin is not easily observable directly by TEM (transmission electron microscope), and is much thinner than that of dinoflagellate.
The difference of the nuclear envelope between Dinoflagellate C. cohnii and that of assembled nuclei from C. cohnii chromosomes in cell-free extracts of Xenopus eggs.
(A) The discontinuous nuclear envelope of dinoflagellate C. cohnii. NE: nuclear envelope. CH: chromosome. Bar=10μm
(B) The continuous nuclear envelope with nuclear pores (arrow indicated) of assembled nuclei. Bar=10m
There is no nuclear assembly in the cell-free system from dinoflagellate
In contrast, nuclear reconstitution did not occur when dinoflagellate C. cohnii chromosomes were incubated with cell-free system from C. cohnii cells. The chromosomes (Fig 5A) were decondensed (Fig 5B) and could not be recondensed to become packed spheres and intact nuclei.
Dinoflagellate C. cohnii chromosomes can not induce nuclear assembly in cell-free extracts from dinoflagellate cells. Being incubated with cell-free extracts from dinoflagellate C. cohnii, the rod-like or dot-like dinoflagellate chromosomes (A) can be decondensed (B), without being recondensed into nucleus-like structure. Samples were stained with the DNA-specific flourescent dye DAPI, and observed with a flourescence microscope at a magnification of 1000 ×.
DISCUSSION
Since Lokha and Maisui first demonstrated that demembranated sperm chromatin can be induced to perform nuclear reassembly in a cell-free system from amphibian eggs components in 19836, a great deal of works followed later suggested that nuclear assembly in vitro can be successfully induced around exogenous DNA or chromatin derived from animals11, plants9 and bacteria7. Thus, nuclear assembly is independent of the species from which DNA was purified. Nuclear assembly is also independent of the length of DNA9, or any specific DNA sequence like genomic DNA or rDNA7,10. Here we showed that exogenously added purified intact chromosomes can be induced to carry out nuclear reconstitution successfully. Our results reinforced the fact that Xenopus egg extracts is a good cell-free system which is extremely stable at room temperature and can be indefinitely stored at −70 °C for studying the assembly of cell nucleus.
The decondensation, recondensation, encapsulation and dispersion of dinoflagellate chromosomes in Xenopus egg extracts suggested that the dinoflagellate chromosome must have been reassembled. It is well known that nucleosome is the basic repeating subunit of typical eukaryotes, which contains a variable length of DNA (≈165-220 bp) and all of the histones. A histone octamer (H2A, H2B, H3, H4) is wrapped by 146bp of DNA in 1.75 super-helical turns in the nucleosome core. In evolutionary terms, histones are among the most highly conserved proteins known. It is surprising, however, that these highly conserved proteins are totally lacking in dinoflagellates and the absence of nucleosomes in dinoflagellate chromatin is due to the absence of histones. This group of unicellular algae must have evolved a different means of packing the nuclear DNA into chromosomes. On the other hand, more than two decades ago, a single histone-like protein has already been reported in dinoflagellate chromatin using one-dimentional gel electrophoresis by Rizzo's group15. Later, an electrophoretic study on C. cohnii using two-dimentional gels showed that a single band in urea gels and SDS gels called HCc16 was actually composed of three similar proteins. These three proteins possess similar biochemical traits just like H4 in higher eukacryotes17. The Xenopus egg extract is a specialized system which possesses two histone variants: histone H2A. X, and histone B4 which the correspond to H2A and H1 histones found in enkaryotic somatic cells. This system is also enriched in the nonhistone protein HMG218. It has been demonstrated that in Xenopus egg extracts naked DNA actually acts as a template for nucleosome assembly19, followed by higher-order chromatin assembly20. Now, in the reconstitution of dinoflagellate chromosomes in Xenopus eggs extracts, the detailed molecular changes involved in the different stages of nuclear assembly, especially the changes of dinoflagellate chromosome DNA and their associated proteins and their interaction with histones and other components in the egg extract, are all not-known. These will be the subjects of our future studies.
Self-assembly is the basic characteristic of many marcromolecular structures in cells. Nuclear reconstitution is also present in the other kinds of cell-free extracts including CHO cells21 and Drosophila melanogaster embryo22. The Xenopus egg is arrested in metaphase of the second meiotic division, which means that stockpiled components (including lamins, pore proteins, and nuclear membrane vesicles) are stored in their disassembled mitotic form until the egg is fertilized or activated. Therefore, the egg extract system contains full components or factors necessary for chromatin decondensation and recondensation to take place in nuclear assembly. In contrast, the dinoflagellate cells extract does not support nuclear reconstitution. We think that this is most probably due to the lack of components, e.g. histones and other necessary factors, in cell-free extracts from dinoflagellate cells. The nuclear envelope of dinoflagellate can be maintained intact during the whole cell cycle3, and chromosomes can be kept condensed permanently1. So there is no necessity to keep a biochemical machine of nuclear assembly and disassembly in dinoflagellate.
References
Soyer MO, Ok Haapala . Division and function of dinoflagellate chromosomes. J Microsc 1974; 19:137–46.
Herzog M, S Von Boleztky, MO Soyer . Ultrastructure and biochemical nuclear aspects of Eukaryote classification: independent evolution of the dinoflagellate as a sister group of the actual eukaryotes? Origin of Life 1984; 13:205–13.
Behaud Y, JM Salmon, MO Soyer . The complex cell cycle of the dinoflagellate protoctist Crythecodinium cohnii as studied in vitro and by cytofluorimentry. J Cell Sci 1991; 100:675–82.
Leoblich AR III .. Dinoflagellate evolution-speculation and evidence. J Protozool 1976; 23:13–28.
Lenaers G, C Scholin, Y Behaud, Saint-Hilaive, M Herzog . A molecular phylogeny of dinoflagellate protists (Pyrrhophyta) inferred from the sequence of 24s rRNA. Divergent domains D1 and D8. J Mol Evol 1991; 32:53–63.
Lokha MJ, Y Maisui . Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science 1983; 220:719–21.
Newport J . Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell 1987; 48:205–17.
Zhang CM, J Qiu, ZH Zhai . Nuclear reconstitution around purified Hela chromosomes in a cell-free system from Xenopus eggs. Acta Biol Exp Sinica 1994; 27:51–9.
Zhang B, Y Chen, ZH Zhai . Plant DNA fragments can induce nuclear reassembly in vitro in Xenopus egg extracts. Science in China 1995; 25:623–30.
Chen Y, B Zhang, XF Li, ZH Zhai . Association of DNA with nuclear matrix in in vitro assembled nuclei induced by rDNA from Tetrahymena shanghaiensis in Xenopus egg extracts. FEBS Letters 1997; 413:449–52.
Qu J, CM Zhang, ZH Zhai . Observation of nuclei reassembled from demembranated Xenopus sperm nuclei and analysis of their lamina components. Cell research 1994; 4:163–72.
Zhang B, Y Chen, ZY Han, H Ris, ZH Zhai . The role of keratin filaments during nuclear envelope reassembly in Xenopus egg extracts. FEBS. Letters 1998; 413:449–52.
Tuttle RC, ARIII Loeblech . An optimal growth medium for the dinoflagellate Crythecodinium cohnii. Phycologia 1975; 14:1–8.
Rizzo PJ . Biochemistry of the dinoflagellate nucleus. In: Talor FJR. eds. The Biology of Dinoflagellates. Blackwell Scientific: Osney Mead, Oxford, England 1987:143–73.
Rizzo PJ, LD Nooden . Chromosomal proteins in the dinoflagellate Gyrodinium C. cohnii. Science 1972; 176:796–7.
Rizzo PJ, RL Morris . Some properties of the histone-like protein from Crythecodinium cohnii (HCc). Bio Systems 1984; 16:211–6.
Vernet G, M Sala-Rovira, M Maeder, F Jacques, M Herzog . Basic nuclear proteins of the histone-less eukaryote Crythecodinium cohnii (Pyrrophyta): two-dimentional electrophoresis and DNA-binding properties. Biochem. Biophys. Acta 1990; 1048:281–9.
Dimitrov S, MC Dasso, AP Wolffe . Remolding sperm chromatin in Xenopus Laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol 1994; 126:591–601.
Laskey RA, AD Mills, NR Morris . Assembly of SV40 chromatin in cell-free system from Xenopus eggs. Cell 1977; 10:237–43.
Hirano T, TJ Mitchison . Cell cycle control of higher-order chromatin assembly around naked DNA in vitro. J Cell Biol 1991; 115:1479–89.
Burke B, L Gerace . A cell-free system to study reassembly of the nuclear envelope at the end of mitosis. Cell 1986; 44:639–52.
Fisher P, M Berrios, G Blobel . Isolation and characterization of a proteinaceous subnuclear fraction composed of nuclear matrix, peripheral lamina, and nuclear pore complexes from embryos of Drosophila melenogaster. J Cell Biol 1982; 92:676–86.
Acknowledgements
We thank Dr Joseph WONG and Prof. Mingxiao DING for providing dinoflagellate Crythecodinium cohnii strain. This work was supported by the National Natural Science Foundation of China (Grant No. 39800075 and 19890380).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
LIU, X., SHEN, Y., CHEN, E. et al. Nuclear assembly of purified Crythecodinium cohnii chromosomes in cell-free extracts of Xenopus laevis eggs. Cell Res 10, 127–137 (2000). https://doi.org/10.1038/sj.cr.7290042
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290042