Abstract
Understanding the local and regional patterns of species distributions has been a major goal of ecological and evolutionary research. The notion that these patterns can be understood through simple quantitative rules is attractive, but while numerous scaling laws exist (e.g., metabolic, fractals), we are aware of no studies that have placed individual traits and community structure together within a genetics based scaling framework. We document the potential for a genetic basis to the scaling of ecological communities, largely based upon our long-term studies of poplars (Populus spp.). The genetic structure and diversity of these foundation species affects riparian ecosystems and determines a much larger community of dependent organisms. Three examples illustrate these ideas. First, there is a strong genetic basis to phytochemistry and tree architecture (both above- and belowground), which can affect diverse organisms and ecosystem processes. Second, empirical studies in the wild show that the local patterns of genetics based community structure scale up to western North America. At multiple spatial scales the arthropod community phenotype is related to the genetic distance among plants that these arthropods depend upon for survival. Third, we suggest that the familiar species–area curve, in which species richness is a function of area, is also a function of genetic diversity. We find that arthropod species richness is closely correlated with the genetic marker diversity and trait variance suggesting a genetic component to these curves. Finally, we discuss how genetic variation can interact with environmental variation to affect community attributes across geographic scales along with conservation implications.
Similar content being viewed by others
Introduction
A general framework for describing patterns in community ecology has proven elusive (Lawton, 1999). Ecological systems are highly complex, and structured by multiple interacting factors whose effects are variable through space and time (Wiens, 1989; Bailey and Whitham, 2006). Simple evolutionary principles are not generally included in explanations of community dynamics or patterns of community structure (e.g., Jones and Lawton, 1995; Hubbell, 2001; Kinzig et al., 2001), and until recently studies of communities have lacked a genetic perspective (but see Dungey et al., 2000; Whitham et al., 2003, 2006; Hochwender and Fritz, 2004; Johnson and Agrawal, 2005; Bangert et al., 2005, 2006a, 2006b; Wimp et al., 2004, 2005; Bailey et al., 2006; Johnson et al., 2006; Tovar-Sánchez and Oyama, 2006; Shuster et al., 2006). Although there has been an explicit incorporation of scale into ecological studies (Wiens, 1989) the integration of genetics and scaling (i.e., genetic scaling; see Table 1 for definitions used throughout this paper) has received little attention (but see Bailey et al., 2004a). In fact, in the special feature of Diversity & Distributions (2006, 12:3): Linking the concept of scale to studies of biological diversity; the word genetics does not occur. We believe that incorporating genetics into scaling is crucial as it advances our evolutionary understanding of scaling relationships.
Incorporating genetics into scaling should further simplify ecological complexity by incorporating evolutionary principles. Ecological scaling has matured over recent decades such that ‘the study of scaling is one powerful way of simplifying ecological complexity and of understanding the physical and biological principles that regulate biodiversity’ (Brown et al., 2002). Although conservation geneticists have considered genetic diversity of a species at the population and species level in order to partition genetic variation using Fst statistics (e.g., Prior et al., 1997), they have not systematically linked these patterns to interactions with other species. At a different scale, Doney et al. (2004) have described oceanic genotypes based on the community metagenome (i.e., the amalgamated genetic composition of co-occurring species) of the microbial community and have discovered novel genes correlated with novel metabolic pathways, which in turn influence large-scale biogeochemical processes. Both of these examples suggest that genetic scaling is an approach that is both feasible and important for understanding patterns in nature.
Because many ecological patterns change with the scale at which they are studied (scale dependence), incorporating scale has proven to be important in understanding patterns such as the species–area relationship (MacArthur and Wilson, 1967). Another familiar example of biological scaling is the way in which metabolism scales with body mass to the ¾ power in endotherms (Savage et al., 2004). Whether ecological relationships are scale dependent or independent, it is important to study these relationships in a genetic context because there is the potential for genetic effects to be important at multiple scales. Ecological patterns that are consistent across scales are of special interest because they may be more easily interpreted. For example, one genetic scaling analysis shows that arthropod community structure is scale independent with plant genetic composition because it scales from local to regional levels (Bangert et al., 2006b).
We suggest that much of ecological scaling is ultimately based upon the fitness consequences of genetic interactions among individuals of different species (Bailey et al., 2006; Shuster et al., 2006). The tendency for these interactions to occur in consistent and repeatable ways can explain emergent properties such as the species–area curve. If scaling ultimately results from intra- and inter-specific interactions between genetically based traits, then scaling can be placed within a genetic and evolutionary framework. Furthermore, when the phenotypic outcomes of individual genotypes are considered as part of relatively simple genetic scaling relationships, traditional quantitative genetics methods can be used to study scaling at population, community, and ecosystem levels (Shuster et al., 2006; Whitham et al., 2003, 2006).
To address this fundamental issue, we focus on the hybridizing complex of Populus spp. as a model system and discuss the relationships between plant genes and community patterns that are both scale dependent and independent. We also provide examples from within single species to show that this concept is not restricted to hybrids. We define genetic scaling as the expression of genes, which affect plant traits, which in turn affects ecological traits (specifically community structure or community phenotypes) at multiple levels of measurement (Table 1). Our definition of genetic scaling is described by the relationship

where S is species richness, c is a normalization constant, and A represents area or the number of trees in a sample, Vg and Ve are genetic and environmental sources of trait variance, the scaling parameter increases at rate m from an initial z0, which is the accumulation scalar. To illustrate how genes scale communities, consider the following example of a population of 20 individuals consisting of three cross types (i.e., two parental species, P1 and P2, and their hybrids, F1). If there are eight P1 each with a trait value of 0, eight P2 with trait values of two and four F1 individuals following an additive expectation with a trait value of 1, then the population would have a mean trait value of 1 and variance equal to 0.84. However, if P2 is completely dominant to P1 (F1 trait value=2), population mean and variance change to 1.2 and 1.01, respectively, illustrating that both a population's trait mean and variance are functions of gene action. If arthropods depend on these trait values, then we would expect arthropod community structure (abundance of each species on the tree population) to scale with the plant population's trait mean (Shuster et al., 2006) and species richness (number of species) to scale with plant population's trait variance (e.g., Wimp et al., 2004; Johnson et al., 2006; Tovar-Sánchez and Oyama, 2006), such that richness in the scenario with P2 dominant would be greater than the scenario where there is just additive gene action. Thus, if we can calculate the mean and variance for a trait as a function of the frequencies of each cross type and gene action (additive, dominance, and so on), we can predict species richness using the genetically based ecological scaling relationship  as previously defined. We suggest that the scaling exponent is a function of phenotypic variation in the plant, and that as genetic (Vg) and environmental (Ve) variation increase, the scaling parameter increases at rate m from an initial z0, that represents the effects of sample size independent of variation.
as previously defined. We suggest that the scaling exponent is a function of phenotypic variation in the plant, and that as genetic (Vg) and environmental (Ve) variation increase, the scaling parameter increases at rate m from an initial z0, that represents the effects of sample size independent of variation.
Evidence that genetic scaling occurs is diverse and we develop different lines of evidence in support of a genetic scaling hypothesis for community structure and species richness. A line cross analysis determines how genes scale traits (additive, dominance, and so on) in a hybridizing species complex from one end of a genetic spectrum to the other. We use this test to show that genetic effects scale important plant traits such as tree architecture and plant phytochemistry. Because these traits affect a diverse community of organisms, the genetic scaling of these traits may ultimately be related to higher levels of organization and provide expectations for a trait variance-species richness relationship. Because foundation species (i.e., species that structure a community by creating locally stable conditions for other species and by modulating and stabilizing fundamental ecosystem processes, Dayton, 1972; Ellison et al., 2005; Table 1), may structure large-scale ecological processes, we examine patterns where community phenotypes are similar at different geographic scales. Finally, we show how species richness in an arthropod community scales as a function of the genetic diversity and trait variation of the trees they depend upon for survival. This genetic-based scaling relationship is an important alternative to the well-known species richness and geographic area scaling relationship. Demonstration of genetic relationships across any of these scales allows us to place community ecology within a scaling and evolutionary framework where currently none exists and testable hypotheses result on the effects of genetic scaling that can be applied to other ecological systems.
Cottonwood system
Cottonwood trees (Populus spp.) are found in most river systems in the western USA where natural intersectional hybridization is common and occurs in an intermediate contact zone. Hybrid zones range in size from a few km to >100 km in length and are characterized by high genetic diversity (Whitham et al., 1999). Populus angustifolia James (narrowleaf cottonwood; section Tacamahaca) hybridizes with P. fremontii Watson (Fremont cottonwood) and P. deltoides Marshall (plains cottonwood), which are both in section Aigeiros. In the Aigeiros × Tacamahaca intersectional hybridizing system, introgression is unidirectional whereby the F1 generation backcrosses only with the narrowleaf parent resulting in a backcross to the narrowleaf generation. Subsequent backcrossing primarily occurs with the narrowleaf parent resulting in a backcross complex (Keim et al., 1989). Our field studies of Populus covered 720 000 km2, were characterized with fAFLPs (fluorescent Amplified Fragment Length Polymorphisms), and were complemented with common garden studies. In 1991, 350 trees representing 81 naturally occurring genotypes of P. fremontii, P. angustifolia, and F1 and backcross hybrids were randomly planted on 4-m centers in a common garden in Ogden, UT, USA (elevation 1370 m; 41°11′N, 111°56′W) and the genetic status of each tree was determined using RFLP (Restriction Fragment Length Polymorphism) markers (Keim et al., 1989; Martinsen et al., 2001).
Scaling across individuals in a population
We first examine the genetic scaling in the Populus hybrid complex using line cross analysis (Lynch and Walsh, 1998; Fritz et al., 2003) of three traits: stem and root fractal architecture, and condensed tannin concentrations. This analysis determines whether traits from parental lineages and their hybrid cross types show recognizable patterns of phenotypic expression in progeny that are consistent with additive and/or dominance effects. For each trait, we have data from four cross types (P. angustifolia, P. fremontii, their F1 hybrids, and backcrosses between F1 and P. angustifolia), which allows us to determine additive and dominant genetic effects of the parental species. We reanalyzed data, with line cross analysis, from Bailey et al. (2004b) for stem fractal (D) architecture and Fischer et al. (2006) for root fractal (D) architecture in an experimental common garden where minirhizotron images (CID-600, CID inc., Vancouver, WA, USA) from Fischer et al. (2006) were analyzed with the same box counting method as for stem architecture (D) in Bailey et al. (2004a). Finally, we reanalyzed data, with line cross analysis, from Schweitzer et al. (2004) in which they determined the percent dry weight concentration of condensed tannins in genotypes from each of the four cross types.
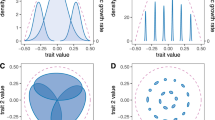
All three of the above phenotypic traits scale across the species complex and plant genetic continuum. These traits are known to affect arthropod communities but the specific genetic effects varied among traits (Table 2). Aboveground architecture was determined by additive gene action (Figure 1a), P. angustifolia was dominant to P. fremontii for belowground root architecture (Figure 1b) and P. fremontii was dominant to P. angustifolia for condensed tannins (Figure 1c). These findings suggest that variation among these traits may be due to different loci underlying their expression or that the traits are driven by different selective pressures. Tree architecture and phytochemistry are each separate links between the tree and the dependent communities. Moreover, stem architecture and tannins are significantly heritable within pure P. angustifolia and each hybrid cross type (Whitham et al., 2006; Bailey et al., 2004a, 2006). The fact that these cottonwood traits genetically scale at two levels of measurement (i.e., within pure species and across the hybrid complex) demonstrates that community and ecosystem patterns are a function of both the within and among cross type genetic variation in this foundation species.
Line cross analysis indicates that genetic effects of hybridizing species vary among traits. Each panel plots the observed mean and SE for each of the three traits: (a) shows that additive genetic effects explain variation in aboveground architecture, (b) shows that additive and significant dominant genetic effects, with narrowleaf dominant to Fremont, explain variation in belowground architecture, and (c) shows that additive and significant dominant genetic effects, with Fremont dominant to narrowleaf, explain variation in foliar condensed tannin concentration.
These findings are also important because they present at least three mechanistic pathways through which plant genetics may scale to other trophic levels; alteration of aboveground stem architecture, belowground root architecture, and phytochemistry. Aboveground architecture has been shown to affect arthropod communities by changing habitat niche space (Morse et al., 1985). Architecture also affects source-sink relationships that affect the fitness of gall-forming insects (Larson and Whitham, 1997), whose presence affects a diverse community of organisms from microbes to birds (Dickson and Whitham, 1996). Similarly, root architecture also varies by plant genotype, which may aid trees in nutrient acquisition and has implications for belowground carbon cycling and belowground communities (Fischer et al., 2006). In addition, phytochemicals such as condensed tannins are known to affect interspecific interactions such as beaver herbivory, avian predation (Bailey et al., 2004a, 2006), and microbial and arthropod community structure (Schweitzer et al., 2006; Whitham et al., 2006; Bangert et al., 2006a). Because each of these plant traits influence dependent community members, a genetic basis to these scaling relationships across individuals may scale up to higher levels by having consequences for the dependent community structure and species richness. However, few studies have attempted to connect the genetic-based patterns at the individual level with biodiversity patterns (but see Vellend, 2005; but see case II of Vellend and Geber, 2005; Shuster et al., 2006), especially at larger spatial scales. Future studies must bridge this gap if we are to realize the potential of genetic scaling to explain many important ecological, evolutionary, and biodiversity patterns.
Community patterns from trees to the region
To address the hypothesis that genetic-based patterns of community phenotypes (e.g., community composition) were robust across geographic scales (i.e., scale independent), we quantified arthropod communities across multiple spatial scales at the individual tree level to the regional level at 720 000 km2. We chose to study the leaf-modifying community (e.g., gall-formers, leaf-rollers, and leaf-tiers) because this community interacts directly with leaf tissue and is likely to be most sensitive to the underlying genetic structure of the plant (Bangert et al., 2006a). In a survey of the arthropods on replicated clones of different cottonwood genotypes, we found that over 57% of the variation among the communities of individual trees within the pure parental species was attributable to heritable differences in their host trees (Shuster et al., 2006). This demonstrates that dependent arthropods can cue into trait differences at the individual tree genotype level at the finest scale of a single population of a pure species. Because individual tree genotype predicts arthropod community phenotypes in a common garden, we predicted that genetic differences among trees at other spatial scales in nature should also predict differences in their community phenotypes.
We extended our common garden study to naturally growing trees and analyzed the relationship between the leaf-modifying arthropod community and cottonwood AFLP genetic composition at multiple spatial scales. We quantified this relationship to address the hypothesis that the relationship between arthropods and plant genetics was qualitatively scale independent and that the genetic effect could be detected from the individual tree to the region. At the tree scale all analyses were on individual trees from riparian zones along six rivers (e.g., St Vrain Creek, CO, USA; Figure 2a). At the stand scale genetic and arthropod compositions were summed across eight trees in each of 10 stands. Likewise, at the river scale the arthropod and AFLP compositions were characterized for each of the six rivers. Finally, at the regional scale we analyzed 137 trees from across the region in a single analysis. Geographic autocorrelation was significant in the regional analysis and was controlled for with a partial Mantel test and the relationship remained largely unchanged indicating that genetic effects were important at each scale (Bangert et al., 2006b).
Arthropod community similarity is negatively correlated with genetic distance among trees at different scales. More closely related trees had more similar arthropod communities. (a) This relationship emerges at the tree scale among 26 individual trees within a single hybrid zone; (b) at the stand scale among 10 stands composed of eight trees each; (c) at the river scale among six rivers; (d) and at the regional scale among 137 trees across 720 000 km2. Panel (d) shows the 9316 points from the Mantel analysis on all pair wise comparisons in gray and the line is fit to the mean distances and similarities for ease of presentation; R2 is reported from the Mantel test. The strength of this relationship declines with increasing scale (e). Note that the scale of the x axis changes with the organizational scale of cottonwoods. (a–d) Modified from Bangert et al., 2006b and P-values from the Mantel tests were derived through a randomization procedure of 9999 randomizations.
Across spatial scales, arthropod community composition was negatively correlated with cottonwood genetic composition at the individual tree, stand, river, and regional scales (Bangert et al., 2006b; Figure 2a–d). These findings show that at all scales examined, more closely related trees (i.e., short genetic distance) support more similar (i.e., high similarity) community phenotypes, whereas more distantly related trees support increasingly different community phenotypes and these differences were not explained by geographic distance.
Moreover, within pure P. fremontii the pattern is still significant (Whitham et al. (2006); R2=0.1852; P<0.001) suggesting that the community response is correlated with fine-scale species-specific genetic composition as well as the larger-scale of genetic differences between two species and their natural hybrids. Preliminary data (GMW, unpublished data) also suggests that the entire free-living arthropod community responds in a similar manner within pure P. fremontii. Furthermore, at a higher scale of analysis involving 23 different genera of trees in central Europe, Brändle and Brandl (2006) found a similar relationship. We defined this genetic scaling relationship as a genetic similarity rule (Bangert et al., 2006a), which shows how a genetic-based community relationship can scale from local to regional levels both within a species and across diverse genera.
Although the genetic similarity rule is scale independent (i.e., it is qualitatively consistent across the local to regional gradient), the strength of the relationship is scale dependent (i.e., it is quantitatively different) and decreases with increasing spatial scale (R2=0.9868, P=0.007; Figure 2e) as predicted by the scale-dependent hypothesis (Johnson and Agrawal, 2005) and the genetic diffusion hypothesis (LeRoy et al., 2006). This suggests that individual differences in trees predict differences in the dependent community at the regional scale with an upper bound on the relationship where environmental variance will likely override the effects of genetic variance.
Implications for the accumulation of biodiversity
Because numerous studies show that diverse species are sensitive to individual plant genotypes, we hypothesized that greater genetic variation in plant traits should lead to greater diversity in the dependent community (Wimp et al., 2004). Furthermore, we expect that particular genetic effects (additive and/or dominance) should also influence the relationship between plant genetic composition and trait variation with species richness. Here we examine how this genetic-based relationship may represent an important, but not necessarily mutually exclusive alternative to the familiar species–area relationship, S=cAz, where species richness is a function of geographic area: S=the number of species, c is a normalization constant, A is the area from which the number of species, S, is quantified, and z is the scaling exponent for the relationship.
We suggest that the species–area relationship, S=cAz, then becomes a species genetic diversity relationship,  , when genetic variance is included as previously described in systems that are strongly driven by bottom-up factors. There is a long history of interpreting the scaling exponent z and the possible mechanisms responsible for its functionality, but previous work has not incorporated genetics. For example, z will differ depending on whether the sample was from an isolated island or the mainland, or whether the community is heavily structured by competition (MacArthur and Wilson, 1967; Strong et al., 1984). However, our work shows that genetic variation in a foundation plant creates habitat variation for dependent species, as our work shows (Bailey et al., 2006; Shuster et al., 2006), and habitat variation predicts species richness (e.g., Vellend, 2005). Consequently, the scaling exponent, z, that relates the species richness of the community dependent on cottonwood trees, should also be related to the genetic variation and diversity among trees.
, when genetic variance is included as previously described in systems that are strongly driven by bottom-up factors. There is a long history of interpreting the scaling exponent z and the possible mechanisms responsible for its functionality, but previous work has not incorporated genetics. For example, z will differ depending on whether the sample was from an isolated island or the mainland, or whether the community is heavily structured by competition (MacArthur and Wilson, 1967; Strong et al., 1984). However, our work shows that genetic variation in a foundation plant creates habitat variation for dependent species, as our work shows (Bailey et al., 2006; Shuster et al., 2006), and habitat variation predicts species richness (e.g., Vellend, 2005). Consequently, the scaling exponent, z, that relates the species richness of the community dependent on cottonwood trees, should also be related to the genetic variation and diversity among trees.
In support of this hypothesis we found a genetic scaling relationship that is remarkably similar to the species–area relationship; as samples are added incrementally (i.e., area increases), species and genetic markers increase with area in a similar manner. We then connect these into a single relationship of species richness as a function of genetic marker accumulation. Area curves (Table 1) were randomly constructed for arthropod species and AFLP markers with 137 trees from across Utah, Colorado, New Mexico, and Arizona, USA (Figure 3a). We then regressed arthropod accumulation against marker accumulation and found that these curves were significantly correlated across the region. When trees were selected at random, arthropods accumulated with AFLP markers (Figure 3b). This is an important relationship because it shows that arthropod species richness may be related to plant genetic diversity and should be considered in conjunction with the species–area relationship.
(a) Scaling in which both arthropods and molecular makers scale similarly as a function of area. At their asymptotes, adequate sampling captures most of the arthropod community and genetic diversity, respectively. (b) Consistent with the above similarity in scaling, there is a strong positive relationship between arthropod species richness and marker richness. Because trees were selected at random, no geographic structure is reflected in these patterns. (c) The species-area scaling exponent (z) is positively related to genetic variation in randomly constructed communities. (d) Species richness is positively correlated with expected trait variance across 12 river systems. P-values in panels (c and d) were derived through a randomization procedure of 9999 randomizations.
From the previous relationship between arthropod and AFLP accumulation it follows that species diversity and richness should be correlated with genetic diversity (Vellend, 2005). Wimp et al. (2004) found that cottonwood gene diversity (He) explained nearly 60% of arthropod community diversity (H′) across 11 stands of trees. Moreover, when they regressed community diversity on stand area, no significant relationship was found. Similarly, Tovar-Sánchez and Oyama (2006) found that genetic diversity at the stand level explained nearly 78% of insect community diversity in a hybridizing oak system across seven oak hybrid zones, independent of area. In combination, these studies in hybrid systems argue that the relationship between animal communities and genetic factors at multiple scales represent an important addition to simple geographic factors such as area or location.
To further investigate the effects of plant genetic variation on dependent community richness while accounting for the effects of geographic area, we re-analyzed data from Wimp et al.'s (2005) common garden experiment. This study quantified the arthropod communities found on 40 trees representing 10 genetically unique individuals from each of four Populus cross types (Fremont, narrowleaf, their F1 hybrids, and F1 × narrowleaf backcrosses). From these data we created 500 communities by randomly sampling a 20-tree subset from the 40-tree pool. For each of the 20-tree communities, we created a species–area curve (see Figure 3a for an example) by determining the mean number of species found on one tree after iteratively sampling random trees, then the mean number of species found on two trees, and so forth up to 20 trees. We found that the species-area scaling parameter, z, increased with increasing genetic variation in the 20-tree subset (Figure 3c). Because this study was conducted in a common garden where environmental effects and area are minimized, this suggests that there is a genetic component to the scalar, z, such that species–area relationships are not simply explained by increasing area and environmental variation.
Finally, we extended our analyses to test the assumption that species richness is related to genetic-based trait variation within a plant population independent of sample size. To evaluate this assumption, we reanalyzed data from Bangert et al. (2005) in which they collected leaf-modifying arthropods from samples of 20 Populus trees found in 12 different river systems. Within each river they determined the proportion of each of three cross types (i.e., Fremont, F1, and the backcross-narrowleaf types using morphology; Wimp et al., 2005). They found that the genotypic differences among rivers predicted community compositional differences (Bangert et al., 2006b). We know from our line-cross analyses that genetic effects vary among traits and we can predict the expected trait variation under different scenarios of genetic effects. We can then relate the genetic effects underlying those traits and trait variation within a river to the species richness found within it. Applying an assumption of additivity to all 12 stands and using the function from before,  , and we solve for Vg by taking the logA of both sides of the equation:
, and we solve for Vg by taking the logA of both sides of the equation:

Thus, we would expect a log-linear relationship between species richness and genetic variation. We then iteratively altered dominance values to identify the degree of dominance that maximized the R2 of the species richness-trait variance relationship.
Using this approach we found that the R2 value was maximized when Fremont genes are almost completely dominant to narrowleaf, and that ∼54% of the differences in species richness among rivers could be explained by differences in expected trait variation (Figure 3d). These findings are consistent with our line cross analysis of tannins (Table 2). In each of the studies illustrated above, genetic scaling of communities occurred. That is, the richness of dependent species partially scaled by the amount of genetic variation present within the plant population's quantitative traits, results that occur when interspecific interactions between individuals depend on heritable traits. We emphasize that these results are based on analyses of genetic structure in a hybridizing species complex in which high levels of genetic variation exist. However, there are two important points that make these patterns general. First, natural hybridization is common and found in diverse taxa worldwide and is thought to represent a major pathway in plant evolution (e.g., Stace, 1987; Smith and Sytsma, 1990; Rieseberg et al., 1996). Second, we have found that other community genetic scaling patterns hold within single species (e.g., P. fremontii and P. angustifolia, Shuster et al., 2006; P. fremontii, Whitham et al., 2006), demonstrating that the scaling relationship holds at the finest level within individual pure species.
Discussion
Scaling of genetic effects
The development of community and ecosystem genetics, that is, ‘the study of genetic interactions among species and their abiotic environment in complex communities’ (e.g., Whitham et al., 2006; but also see Antonovics, 1992; Whitham et al., 2003; Johnson and Agrawal, 2005; Shuster et al., 2006), reveals the importance of understanding whether genetic interactions among species at local scales also predict the associations of plant genes and dependent species at regional scales (Bangert et al., 2005, 2006b). Importantly, if the underlying genetic basis of community-level scaling rules can be elucidated, then ecological scaling can be evaluated within an evolutionary framework.
The findings of previous studies on the subject of community scaling are mixed. For example, only a few studies include genetic composition, and these only cover small spatial scales, which prevents an evaluation of genetic scaling at larger geographic scales. However, in a study of the evening primrose (Oenothera biennis), Johnson and Agrawal (2005) found that plant genotype affected ecological measures of arthropod community structure at a small scale, whereas environment was more important at slightly larger scales, thus showing scale-dependent patterns. They found strong environment and gene × environment interaction (G × E) effects for several arthropod community parameters suggesting that arthropod communities respond more strongly to different plant genotypes based on environmental context. Several other studies have concluded that plant genotype effects were weak and that the environment was most responsible for arthropod population and community structure at the scales of these studies (a few meter to a few kilometer; Maddox and Cappuccino, 1986; Stiling and Rossi, 1995, 1996; Stiling and Bowdish, 2000; Johnson and Agrawal, 2005). Interestingly, the studies that did not demonstrate genetic scaling, also did not involve foundation species. This result is consistent with our hypothesis that genetic scaling is most likely to occur with foundation species that have large effects on the community and ecosystem (see Ellison et al., 2005).
Our studies of a foundation species also show that genetic effects associated with individual tree genotypes scale to higher trophic levels (Bailey et al., 2006), a conclusion supported by independent lines of investigation. There is heritable variation in the scaling relationship of whole tree architecture (Bailey et al., 2004b) and architecture has been shown to affect arthropods in several studies (e.g., Morse et al., 1985; Larson and Whitham, 1997). When community structure is examined across multiple scales community patterns remain qualitatively constant (a few meters to 720 000 km2; Bangert et al., 2005, 2006b). Finally, we show that species–area curves may have a genetic basis and are not strictly governed by area. These findings are important because they represent an initial first examination of genetic scaling of ecological communities.
We emphasize that different spatial patterns of genetic scaling are not mutually exclusive. For example, Thompson's geographic mosaic hypothesis of coevolution (2005) argues that because species interactions vary geographically, different populations of plants may exhibit different patterns with the dependent community across the landscape. Thus, the geographic mosaic could affect evolutionary trajectories of communities at different geographic localities and scales. However, while the specific interactions among species may change geographically the relationship of species diversity with the underling genetic diversity of the foundation species may remain relatively constant over a wide range of scales (Bangert et al., 2006b). In other words, while local communities may have different evolutionary trajectories, fundamental relationships of species diversity with genetic diversity may be scale independent. Although the appropriate data for addressing which genetic relationships are either scale dependent or independent are currently unavailable, it is important for future studies to develop a predictive theory and empirically address this issue for both pure and hybridizing species.
Are these patterns specific to hybrids?
No. Although many of our initial studies in community genetics involved hybrid cottonwoods, subsequent studies within single species exhibited similar patterns. For example, initial studies of the relationship between genetic distance and arthropod community similarity involved cottonwood parentals and their hybrids, additional studies became more refined and considered the hybrids only, and finally considered P. fremontii only (Bangert et al., 2006a, 2006b). Other studies have shown significant heritability for multiple community traits across multiple trophic levels within both pure species and hybrids (Bailey et al., 2006; Shuster et al., 2006; Whitham et al., 2006). For example, Shuster et al. (see Tables 1 and 2 in Shuster et al., 2006) found that 57% of community phenotype variation was due to the genetic differences among genotypes within either pure P. fremontii or P. angustifolia, which was 3 × greater than community variation between these two species and their hybrids (19%). Moreover, many community metrics scale across the plant taxonomic hierarchy at large geographic scales from genera to the species level (e.g., genera: Brändle and Brandl, 2006; hybrids: Wimp et al., 2004; Tovar-Sánchez and Oyama, 2006; Bangert et al., 2006b; species: Whitham et al., 2006). Thus, genetic scaling has been detected in both phylogenetic and spatial analyses. Although hybrids are associated with high genetic variability, the genetic variability within single species is sufficient to drive genetic scaling at this finer level, which increases the potential for genetic scaling to be widespread.
Predicting genetic scaling
In light of previous findings that genetic effects are thought to be important or detectable only at small spatial scales (scale-dependent hypothesis; Johnson and Agrawal, 2005), we suggest that hypotheses need to be developed to predict when genes will scale to large spatial scales and when they will not in both pure and hybridizing species. Several factors may affect genetic scaling of communities. First, community genetic scaling over geographic space is likely determined by the relative contributions of genetic variation found within and among interacting species and the environmental variation surrounding those interactions (Vellend, 2005; Shuster et al., 2006). For example, naturally hybridizing plant systems tend to be associated with high genetic variation owing to recombination of parental species genomes (Rieseberg and Ellstrand, 1993; Whitham et al., 1999; Dungey et al., 2000; Hochwender and Fritz, 2004), whereas non-hybridizing species may have relatively less genetic variation. However, pure foundation species are often widespread and are known to exhibit high genetic variation (e.g., aspen; Mitton and Grant, 1996). In an experiment with the foundation species, Zostera marina a common sea grass, Reusch et al. (2005) found that plots with higher genetic diversity increased resistance to extreme heat events and positively benefited the associated invertebrate fauna. Likewise, in another experiment with a foundation species Crutsinger et al. (2006) found that as the genetic diversity of Solidago altissima increased both arthropod species richness and annual net primary production also increased. The studies to date on foundation species suggest the intraspecific genetic diversity can influence the associated community and ecosystem processes, it is important to extend these studies to multiple spatial scales.
Second, genetic traits in foundation species should be far more likely to scale than genetic traits in non-foundation species, and least of all in rare species. This simply reflects their relative importance in structuring communities and affecting ecosystem-level processes such as nutrient cycling (Ellison et al., 2005). In a survey of eight different study systems where strong genetic effects on the community had been documented, Whitham et al. (2006) found that all but one of the systems could be considered a foundation species. The importance of a trait at population, community, and regional levels is dependent upon the variability of that trait at each scale. Third, traits like condensed tannins (Whitham et al., 2003, 2006) that produce community and ecosystem phenotypes would also be likely to scale. Thus, genetic scaling should be most likely to occur with genes that result in large phenotypic effects that are expressed in foundation species.
We present a conceptual model to illustrate these points. As geographic scale increases, it is reasonable to assume that environmental variability increases. If genetic variation in one or more of the interacting species also increases at a similar rate, then community genetic effects should be maintained, and we are most likely to find that genetic differences among individuals predict differences in communities regardless of geographic scale (see Vellend, 2003 for genetic diversity × species diversity relationships by area). Conversely, if the variability of genetic effects does not increase as fast as environmental variation, then the environment becomes a stronger organizing factor on dependent communities and the genetic effects on the community decline. Johnson and Agrawal's (2005) study of a non-foundation species is a good empirical example consistent with this model. They found that environmental effects dominated their results, as plant genetic variation remained constant across five different environments. In contrast, our studies of a foundation species showed significant genetic scaling from local to regional levels (Figure 2). In combination, these studies provide the beginnings of a predictive framework for when genes should and should not scale.
Conservation and genetic scaling
Traditional scaling methods for identifying patterns of species distribution and diversity are based upon scale-dependent metrics that are often limited in their ability to describe community patterns and have been criticized (Strong et al., 1984). However, Calder (2000) advocates the use of scaling to cautiously inform conservation decisions. For example, if the species–area relationship is more appropriately a species genetic diversity relationship as Figure 3b suggests, then greater focus should be placed on maintaining the genetic diversity of both foundation species along with the geographic areas they occupy.
Studies in five different systems found that plots with the highest plant genetic diversity supported significantly greater invertebrate species richness than plots with low genetic diversity in both hybridizing and single species systems (Wimp et al., 2004; Reusch et al., 2005; Crutsinger et al., 2006; Johnson et al., 2006; Tovar-Sánchez and Oyama, 2006). Moreover, analyses of arthropod communities in the wild and in common gardens show that many arthropod species are tree genotype dependent (McIntyre and Whitham, 2003; Wimp et al., 2005). Thus, maximizing plant genetic diversity maximizes species diversity. These same patterns were found over a broad geographic area. These findings argue that genetic diversity in common species is important for maintaining biodiversity at regional scales (Bangert et al., 2005). Collaborative restoration studies with Reclamation using P. fremontii collected from across the western USA are currently ongoing to study the genetic scaling implications for the dependent community.
Conclusions
Community ecology has struggled to find predictive models and a unifying theoretical framework that can be applied across multiple systems and scales (Lawton, 1999). We have demonstrated the potential for genetic factors to provide a predictive model for community structure that may be applied across systems and scale from individuals to geographic regions. Approaching community ecology from a genetic perspective also allows us to incorporate evolutionary principles, e.g., natural selection, into conserving species, species interactions, and their evolutionary potential. Consequently, evolution, population genetics, and community ecology can be theoretically united (Vellend, 2005).
References
Antonovics J (1992). Toward community genetics. In: Fritz RS, Simms EL (eds). Plant Resistance to Herbivores and Pathogens. University of Chicago Press: Chicago. pp 426–449.
Bailey JK, Whitham TG (2006). Biodiversity is related to indirect interactions among species of large effect. In: Ohgushi T, Craig T, Price PW (eds). Indirect Interaction Webs: Nontrophic Linkages Through Induced Plant Traits. Cambridge University Press: Cambridge (in press).
Bailey JK, Bangert RK, Schweitzer JA, Trotter RT, Shuster SM, Whitham TG (2004a). Fractal geometry is heritable in trees. Evolution 58: 2100–2102.
Bailey JK, Schweitzer JA, Rehill BJ, Lindroth RL, Martinsen GD, Whitham TG (2004b). Beavers as molecular geneticists: a genetic basis to the foraging of an ecosystem engineer. Ecology 85: 603–608.
Bailey JK, Wooley SC, Lindroth RL, Whitham TG (2006). Importance of species interactions to community heritability: a genetic basis to trophic level interactions. Ecol Lett 9: 78–85.
Bangert RK, Allan GJ, Turek RJ, Wimp GM, Meneses N, Martinsen GD et al. (2006b). From genes to geography: a genetic similarity rule for arthropod community structure at multiple geographic scales. Mol Ecol (in press).
Bangert RK, Turek RJ, Martinsen GD, Wimp GM, Bailey JK, Whitham TG (2005). Benefits of conservation of plant genetic diversity on arthropod diversity. Cons Biol 19: 379–390.
Bangert RK, Turek RJ, Rehill B, Allan GJ, Wimp GM, Schweitzer JA et al. (2006a). A genetic similarity rule determines arthropod community structure. Mol Ecol 15: 1379–1392.
Brändle M, Brandl R (2006). Is the composition of phytophagous insects and parasitic fungi among trees predictable? Oikos 113: 296–304.
Brown JH, Gupta VK, Li B-L, Milne BT, Restrepo C, West GB (2002). The fractal nature of nature: power laws, ecological complexity and biodiversity. Philos Trans R Soc Lond B 357: 619–626.
Calder WA (2000). Diversity and convergence: scaling for conservation. In: Brown JH, West GB (eds). Scaling in Biology. Oxford University Press: New York. pp 297–323.
Crutsinger GM, Collins MD, Fordyce JA, Gompert Z, Nice CC, Sanders NJ (2006). Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313: 966–968.
Dayton PK (1972). Toward an understanding of community resilience and the potential effects of enrichments to the benthos at McMurdo Sound, Antarctica. In: Parker BC (ed). Proceedings of the Colloquium on Conservation Problems in Antarctica. Allen Press: Lawrence. pp 81–96.
Dickson LL, Whitham TG (1996). Genetically-based plant resistance traits affect arthropods, fungi, and birds. Oecologia 106: 400–406.
Doney SC, Abbott MR, Cullen JJ, Karl DM, Rothstein L (2004). From genes to ecosystems, the ocean's new frontier. Front Ecol Environ 2: 457–466.
Dungey HS, Potts BM, Whitham TG, Li HF (2000). Plant genetics affects arthropod community richness and composition: evidence from a synthetic eucalypt hybrid population. Evolution 54: 1938–1946.
Ellison AM, Bank MS, Clinton BD, Colburn EA, Elliott K, Ford CR et al. (2005). Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front Ecol Environ 3: 479–486.
Fischer DG, Hart SC, Rehill BJ, Lindroth RL, Keim P, Whitham TG (2006). Do high tannin leaves require more roots? Oecologia 149: 668–675.
Fritz RS, Hochwender CG, Brunsfeld SJ, Roche BM (2003). Genetic architecture of susceptibility to herbivores in hybrid willows. J Evol Biol 16: 1115–1126.
Hochwender CG, Fritz RS (2004). Plant genetic differences influence herbivore community structure: evidence from a hybrid willow system. Oecologia 138: 547–557.
Hubbell SP (2001). The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press: Princeton.
Johnson MTJ, Agrawal AA (2005). Plant genotype and environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis). Ecology 86: 874–885.
Johnson MTJ, Lajeunesse MJ, Agrawal AA (2006). Additive and interactive effects of plant genotypic diversity on arthropod communities and plant fitness. Ecol Lett 9: 24–34.
Jones CG, Lawton JH (eds) (1995). Linking Species and Ecosystems. Chapman & Hall: New York.
Keim P, Paige KN, Whitham TG, Lark KG (1989). Genetic analysis of an interspecific hybrid swarm of populus: occurrence of unidirectional introgression. Genetics 123: 557–565.
Kinzig AP, Pacala SW, Tilman D (eds) (2001). The Functional Consequences of Biodiversity. Princeton University Press: Princeton.
Larson KC, Whitham TG (1997). Competition between gall aphids and natural plant sinks: plant architecture affects resistance to galling. Oecologia 109: 575–582.
Lawton JH (1999). Are there general laws in ecology? Oikos 84: 177–192.
Legendre P, Legendre L (1998). Numerical Ecology, 2nd edn. Elsevier: Amsterdam.
LeRoy CJ, Whitham TG, Keim P, Marks JC (2006). Plant genes link forests and streams. Ecology 87: 255–261.
Lynch M, Walsh B (1998). Genetics and Analysis of Quantitative Traits. Sinauer: Sunderland.
MacArthur RH, Wilson EO (1967). The Theory of Island Biogeography. Princeton University Press: Princeton.
Maddox GD, Cappuccino N (1986). Genetic determination of plant susceptibility to an herbivorous insect depends on environmental context. Evolution 40: 863–866.
Magurran AE (1988). Ecological Diversity and its Measurement. Princeton University Press: Princeton.
Martinsen GD, Whitham TG, Turek RJ, Keim P (2001). Hybrid populations selectively filter gene introgression between species. Evolution 55: 1325–1335.
McIntyre PJ, Whitham TG (2003). Plant genotype affects long-term herbivore population dynamics and extinction: conservation implications. Ecology 84: 311–322.
Mitton JB, Grant MC (1996). Genetic variation and the natural history of quaking aspen. Bioscience 46: 75–81.
Molles Jr MC (1999). Ecology. McGraw-Hill: Boston.
Morse DR, Lawton JH, Dodson MM, Williamson MH (1985). Fractal dimension of vegetation and the distribution of arthropod body lengths. Nature 314: 731–733.
Prior KA, Gibbs HL, Weatherhead PJ (1997). Population genetic structure in the black rat snake: implications for management. Cons Biol 11: 1147–1158.
Reusch TBH, Ehlers A, Hämmerli A, Worm B (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc Natl Acad Sci USA 102: 2826–2831.
Rieseberg LH, Ellstrand NC (1993). What can molecular markers tell us about plant hybridization? Crit Rev Plant Sci 12: 213–241.
Rieseberg LH, Sinervo B, Linder CR, Ungerer MC, Arias DM (1996). Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272: 741–745.
Savage VM, Gillooly JF, Woodruff WH, West GB, Allen AP, Enquist BJ et al. (2004). The predominance of quarter-power scaling in biology. Funct Ecol 18: 257–282.
Schweitzer JA, Bailey JK, Bangert RK, Hart SC, Whitham TG (2006). The role of plant genetics in determining above- and belowground microbial communities. In: Phyllosphere Microbiology. APS Press: St Paul (in press).
Schweitzer JA, Bailey JK, Rehill BJ, Martinsen GD, Hart SC, Lindroth RL et al. (2004). Genetically based trait in a dominant tree affects ecosystem processes. Ecol Lett 7: 127–134.
Shuster SM, Lonsdorf EV, Wimp GM, Bailey JK, Whitham TG (2006). Community heritability measures the evolutionary consequences of indirect genetic effects on community structure. Evolution 60: 991–1003.
Smith RL, Sytsma KJ (1990). Evolution of Populus nigra (sect. Aigeiros): introgressive hybridization and the chloroplast contribution of Populus alba (sect. Populus). Amer Jour Bot 77: 1176–1187.
Stace CA (1987). Hybridization and the plant species. In: Urbanska KM (ed). Differential Patterns in Higher Plants. Academic Press: New York. pp 115–127.
Stiling P, Bowdish TI (2000). Direct and indirect effects of plant clone and local environment on herbivore abundance. Ecology 81: 281–285.
Stiling P, Rossi AM (1995). Coastal insect herbivore communities are affected more by local environmental conditions than by plant genotype. Ecol Entomol 20: 184–190.
Stiling P, Rossi AM (1996). Complex effects of genotype and environment on insect herbivores and their enemies. Ecology 77: 2212–2218.
Strong Jr DR, Simberloff D, Abele LG, Thistle AB (eds) (1984). Ecological Communities: Conceptual Issues and the Evidence. Princeton University Press: Princeton.
Thompson JN (2005). The Geographic Mosaic of Coevolution. University of Chicago Press: Chicago.
Tovar-Sánchez E, Oyama K (2006). Effect of hybridization of the quercus crasssifolia × quercus crassipes complex on the community structure of endophagous insects. Oecologia 147: 702–713.
Vellend M (2003). Island biogeography of genes and species. Am Nat 162: 358–365.
Vellend M (2005). Species diversity and genetic diversity: parallel processes and correlated patterns. Am Nat 166: 199–215.
Vellend M, Geber MA (2005). Connections between species diversity and genetic diversity. Ecol Lett 8: 767–781.
Whitham TG, Bailey JK, Schweitzer JA, Shuster SM, Bangert RK, LeRoy CJ et al. (2006). Community and ecosystem genetics: a framework for integrating from genes to ecosystems. Nat Rev Genet 7: 510–523.
Whitham TG, Martinsen GD, Floate KD, Dungey HS, Potts BM, Keim P (1999). Plant hybrid zones affect biodiversity: tools for a genetic-based understanding of community structure. Ecology 80: 416–428.
Whitham TG, Young WP, Martinsen GD, Gehring CA, Schweitzer JA, Shuster SM et al. (2003). Community and ecosystem genetics: a consequence of the extended phenotype. Ecology 84: 559–573.
Wiens JA (1989). Spatial scaling in ecology. Funct Ecol 3: 385–397.
Wiens JA (2001). Understanding the problem of scale in experimental ecology. In: Gardener RH, Kemp WM, Kennedy VS, Petersen JE (eds). Scaling Relations in Experimental Ecology. Columbia University Press: New York. pp 61–88.
Wimp GM, Martinsen GD, Floate KD, Bangert RK, Whitham TG (2005). Plant genetic determinants of arthropod community structure and diversity. Evolution 59: 61–69.
Wimp GM, Young WP, Woolbright SA, Martinsen GD, Keim P, Whitham TG (2004). Conserving plant genetic diversity for dependent animal communities. Ecol Lett 7: 776–780.
Acknowledgements
We thank the Ogden Nature Center, the Utah Department of Natural Resources, Cibola National Wildlife Refuge, and Reclamation for supporting our restoration and common garden studies. Discussions with CJ LeRoy and the cottonwood ecology group helped to clarify the ideas presented in this manuscript. Funding was provided by NSF Grants DEB-0078280, DEB-0425908, Reclamation, and the Merriam-Powell Center for Environmental Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bangert, R., Lonsdorf, E., Wimp, G. et al. Genetic structure of a foundation species: scaling community phenotypes from the individual to the region. Heredity 100, 121–131 (2008). https://doi.org/10.1038/sj.hdy.6800914
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800914
Keywords
This article is cited by
-
Experimental drought reduces genetic diversity in the grassland foundation species Bouteloua eriopoda
Oecologia (2019)
-
Hybridization between Tithonia tubaeformis and T. rotundifolia (Asteraceae) evidenced by nSSR and secondary metabolites
Plant Systematics and Evolution (2018)
-
Genetic diversity affects ecological performance and stress response of marine diatom populations
The ISME Journal (2016)
-
Effect of host-plant genetic diversity on oak canopy arthropod community structure in central Mexico
Revista Chilena de Historia Natural (2015)
-
Oak canopy arthropod communities: which factors shape its structure?
Revista Chilena de Historia Natural (2015)