Abstract
All over Europe, the number of draught horses has decreased drastically during the last 50 years. As a prerequisite for efficient management decisions, we analysed the conservation status in Austrian (Noriker Carinthia – NC, Noriker Salzburg – NS), Croatian (Croatian Coldblood – C, Posavina horse – P) and German (Altmaerkisch Coldblood – A, Black Forest horse – BF, Mecklenburg Coldblood – M, Rhenish German Draught horse – R, Saxon Thuringa Coldblood – ST, Schleswig Draught horse – Sch, South German Coldblood – SG) draught horses (434) using multilocus genotypic information from 30 (effectively 27) microsatellite loci. Populations located in areas with less intensive agricultural production (C, NC, NS, P and SG) had greater diversity within the population and estimated effective population size than A, BF, Sch, M, R and ST populations. The PCA plots revealed that populations form five separate groups. The ‘Noriker’ group (NC, NS and SG) and the ‘Rhenish’ group (A, M, R and ST) were the most distinctive (pairwise FST values ranged from 0.078 to 0.094). The ‘Croatian’ group (C and P) was in the centre, while the BF and Sch populations formed two out-groups. A posterior Bayesian analysis detected further differentiation, mainly caused by political and geographical factors. Thus, it was possible to separate the South German Coldblood from the Austrian Noriker population where no subpopulation structure was detected. The admixture analysis revealed imprecise classification between C and P populations. A small but notable separation of R from A, M and ST populations was detected, while Sch and BF populations remained as out-groups. The information obtained should aid in making efficient conservation decisions.
Similar content being viewed by others
Introduction
The conservation of draught horses is of increasing concern, because apart from their historical and genetic value (see Hall and Bradley, 1995), they are still directly connected to agricultural production with extensive and traditional farm management. Moreover, they are often used in agriculturally harsh areas like mountain pastures, semi-deserts and swamps, where they are a part of very specific and rare ecosystems (Miraglia et al., 2006). Unfortunately, these highly specialized horses are threatened with extinction, especially in agriculturally to developed European countries. A drastic example is the geographical area of the former Austro-Hungarian Monarchy and Germany where, in 1876, about 4 000 000 different purebred draught horses and heavy crossbreed animals (Josche, 1878) existed. Nowadays, in the same geographical area, there remain only about 20 000 draught horses.
Here, we are referring to 11 European draught horse populations from Austria (Noriker Carinthia – NC, Noriker Salzburg – NS), Croatia (Posavina horse – P, Croatian Coldblood – C) and Germany (Altmaerkisch Coldblood – A, Black Forest horse – BF, Mecklenburg Coldblood – M, Rhenish German Draught horse – R, Saxon Thuringa Coldblood – ST, Schleswig Draught horse – Sch, South German Coldblood – SG).
The two Austrian Noriker populations (Carinthia and Salzburg) are considered as one breed kept by two distinct breeding organisations, geographically separated by the Alps. The South German Coldblood horse, a daughter population of the Noriker horse, is kept mainly in Bavaria, a region in Germany that is geographically and historically closely related to Austria. The Black Forest horse has its origin in the Black Forest and its main distribution is in the Baden-Württemberg region. Although considered to be related to Noriker horses, Aberle et al. (2004) have shown recently that the Black Forest horse is quite distinct from the South German Coldblood. In Croatia, two main draught horse populations, the Croatian Coldblood and the Posavina horse, are intermixed. Several factors have led to the admixture of these two populations. The recent war in Croatia created a situation where a substantial number of pedigree records and breeding animals were lost or changed identity. After the war, new herd books and administration networks were re-established, assigning breed membership according to phenotypic appearance. Furthermore, the export of horse meat is currently the main income of breeders; hence the smaller autochthonous Posavina type will likely be crossed with the larger Croatian Coldblood. Altmaerkisch, Mecklenburg, Rhenish and Saxon Thuringa draught horses were once considered to be four populations of one breed, founded by breeding primarily with Belgian Draught horses. Later, Altmaerkisch, Mecklenburg and Saxon Thuringa were politically separated from the Rhenish population. The Schleswig Draught horse is bred in northern Germany (mainly in Schleswig-Holstein and Lower Saxony) and is most closely related to the neighbouring Jutland horse from Denmark.
The general objective of this study was to infer the recent genetic relationship among breeds and the population structure of indigenous draught horse populations from Austria, Croatia and Germany. The study was based on microsatellite markers, a posterior Bayesian approach using the STRUCTURE program (Pritchard et al., 2000; Falush et al., 2003) and various multivariate techniques. Analyses of microsatellite information with a posterior Bayesian approach (Glowatzski-Mullis et al., 2006) or/and other multivariate techniques (Achmann et al., 2004; Cothran and Luis, 2005) have already proven to be a powerful means of resolving subtle conservation genetic issues in horse populations.
In addition, we wanted to provide answers to specific hypotheses related to the population or/and breed membership and to future conservation management decisions.
Methods
Sampling and laboratory techniques
Blood or hair samples were randomly taken from 11 populations located from the Baltic to the Adriatic Sea, over the central European area (see Figure 1). A total of 434 individual horse samples were taken, originating from three countries: (i) Austria: Noriker Carinthia (45) and Noriker Salzburg (45); (ii) Croatia: Croatian Coldblood (41) and Posavina horse (45) and (iii) Germany: Altmaerkisch Coldblood (32), Black Forest horse (45), Mecklenburg Coldblood (22), Rhenish German Draught horses (46), Saxon Thuringa Coldblood (23), Schleswig Draught horse (45) and South German Coldblood (45). We avoided intentional sampling of closely related animals. For Croatian populations, no accurate pedigree information is available and the samples were taken randomly from 20 distinct herds. It is the opinion of the authors that this sufficiently represents the sampled population. Aberle et al. (2004) used pedigree information as a basis for taking the blood/hair samples from German populations. Genomic DNA was extracted from whole blood using the QIAamp 96 DNA Blood Kit (Qiagen, Hilden, Germany), and from hair root samples using the DNeasy Tissue Kit (Qiagen), following the manufacturer's protocol. Amplification and molecular analysis of the 30 microsatellite markers (see Table 1) were the same as described by Aberle et al. (2004).
Genetic diversity, population differentiation and relationship
Allele frequency, number of alleles, observed heterozygosity and gene diversity (heterozygosity expected from Hardy–Weinberg equilibrium assumptions) were calculated across different loci and/or populations (breeds) using GENETIX 4.05.2 (Belkhir et al., 1996–2004; http://www.univ-montp2.fr/~genetix/genetix/intro.htm). The mean number of alleles (MNA) and rarefacted mean numbers of alleles (RMNA) were calculated using MOLKIN 2.0 (Gutiérrez et al., 2005; http://www.ucm.es/info/prodanim/Molkin2.zip). Tests for deviations from Hardy–Weinberg equilibrium (HWE) were performed across all loci by GENEPOP 3.4 (Raymond and Rousset, 1995; ftp://ftp.cefe.cnrsmop.fr/genepop), applying the exact test with default settings of the Markov chain Monte Carlo methodology (Guo and Thompson, 1992). Where appropriate, P-values were adjusted according to Hochberg's step-up Bonferroni method (Hochberg, 1988) using the SAS package (SAS Institute, 1999–2001). Wright's FIS was calculated according to Weir and Cockerham (1984) using GENETIX 4.05.2. Effective population size (Ne) was calculated with NeESTIMATOR 1.3 (Peel et al., 2004; http://www.dpi.qld.gov.au/fishweb/11674.html) using linkage (gametic) disequilibrium method presented by Hill (1981).
Molecular genetic relationships among populations were derived using Wright's FST (Weir and Cockerham, 1984), molecular kinship distance Dk (Caballero and Toro, 2002) and Slatkin's RST (Slatkin, 1995). These three measures of population differentiation are intended to provide different information. FST is not based on an evolutionary model and has been shown to be appropriate for the analysis of closely related populations. Molecular kinship distance (Dk) accounts for the allele frequencies in the founder populations (Alvarez et al., 2005). RST is based on the mutation rate of marker length under the assumption of a strict stepwise mutation model (SMM). Population differentiation measures FST, Dk and RST were calculated using GENETIX 4.05.2, MOLKIN 2.0 and SPAGeDI (Hardy and Vekemans, 2002; http://www.ulb.ac.be/sciences/ecoevol/spagedi.html), respectively. Furthermore, three matrices showing differentiation among populations (based on FST, Dk or RST) were used for principal component analyses (PCA). PCA graphic illustrations were generated by the SAS software (SAS Institute, 1999–2001).
Individual-based analysis of population structure and admixture level
Population structure was inferred using STRUCTURE 2.1 (Pritchard et al., 2000, http://www.pritch.bsd.uchicago.edu/software/structure2_1.html). The program enables estimation of a ‘hidden structure’, that is the number of different clusters (K partitions) obtained without using any a priori information about individual membership (population and/or breed). Furthermore, the program is able to determine the corresponding fraction of an individual's genome derived from an ancestry in one of the clusters (K) determined by the program. The program STRUCTURE uses the Markov chain Monte Carlo (MCMC) method, see Pritchard et al. (2000) and Falush et al. (2003), and estimates the natural logarithm of the probability (Pr) of the observed genotypic array (G), given a preassigned number of clusters (parameter K) in the dataset [ln Pr(G∣K)]. In a Bayesian set-up the estimate of ln Pr(G∣K) is a direct indicator of the posterior probability of having K number of clusters, given the observed genotypic array (G). To obtain a representative value of K for modelling the data, we ran 10 independent runs of the Gibbs sampler for each K between 1 and 11 with a burn-in length of 105 followed by 106 iterations. In all runs we used default settings, that is, an admixture model with correlated frequencies and the parameter of individual admixture alpha set to be the same for all clusters and with a uniform prior. The analysis performed on the whole data set was also repeated for reduced data sets considering only the ‘Rhenish’ group (Altmaerkisch Coldblood, Mecklenburg Coldblood, Rhenish German Draught horse and Saxon Thuringa Coldblood), the ‘Noriker’ group (Noriker-Carinthia, Noriker-Salzburg and South German Coldblood) and the ‘Croatian’ group (Croatian Coldblood and Posavina horse). The resulting individual genotype membership coefficients were displayed using DISTRUCT (Rosenberg, 2004, http://rosenberglab.bioinformatics.med.umich.edu/distruct.html). For a more detailed description about the Bayesian approach implemented in STRUCTURE see Pearse and Crandall (2004) and Chikhi and Bruford (2005).
Results
Diversity and characteristics of microsatellite loci
A description of the markers used, including chromosome localisation, size range, number of alleles per marker, observed heterozygosity, FST and RST values, is given in Table 1. The total number of alleles found for the 30 microsatellite markers in 11 draught horse populations was 270. The mean number of alleles per locus was 9, with a range of 6–15. Observed heterozygosity ranged from 0.291 for locus HTG07 to 0.837 for locus ASB17. After Hochberg's step-up Bonferroni correction for multiple comparisons (Hochberg, 1988), a significant deviation from HWE was observed for loci COR056, COR071 and LEX007. Although the existence of null alleles was not proven, those three loci were excluded from further analyses.
Genetic diversity within populations
Genetic diversity within 11 draught horse populations is presented in Table 2. The lowest observed heterozygosity (0.645) and gene diversity (0.649) were present in Mecklenburg Coldblood, while the highest observed heterozygosity (0.722) and gene diversity (0.706) were present in Croatian Coldblood. The lowest allele richness was present in Saxon Thuringa Coldblood (MNA=5.30) and the highest in Croatian Coldblood (MNA=7.11). However, the mean number of alleles is influenced by sample size, and when adjusted to sample size (42 alleles), the lowest allele richness was present in the Schleswig Draught horse (RMNA=5.29), while Croatian Coldblood still had the highest allele richness (6.68). Negative FIS coefficients, confidence interval (95%) ranging from −0.107 to −0.017, were estimated only for Saxon Thuringa Coldblood, suggesting an excess of heterozygotes due to non-random mating. In all other populations, the confidence intervals (95%) for FIS coefficients ranged from negative to positive values. Estimated effective population size was lowest in Mecklenburg Coldblood (31.6) and highest in Noriker-Salzburg (164.0).
In general, the above-average diversity values (heterozygosity, allele richness and effective population size) were observed in Southern populations (Croatian Coldblood, Noriker-Carinthia, Noriker-Salzburg, Posavina horse and South German Coldblood).
Genetic differentiation and relationship between populations
Differentiation, FST values, among the 11 draught horse populations is shown in Table 3. The largest differentiation was observed between the Black Forest horse and the Schleswig Draught horse (10.6% of the total variability; FST=0.106). Small differentiation, from FST=0.012 to FST=0.041, was observed within populations once considered to be Rhenish German Coldblood (Altmaerkisch Coldblood, Mecklenburg Coldblood, Rhenish German Draught horse and Saxon Thuringa Coldblood), then from FST=0.005 to FST=0.031 within Noriker horses (Noriker-Carinthia, Noriker-Salzburg and South German Coldblood), and FST=0.018 between Croatian Coldblood and the Posavina horse.
Three-dimensional PCA plots illustrate the molecular genetic relationship among the 11 draught horse populations (Figure 2). No matter upon which matrix the principal component analysis was based (FST, Dk or RST), a large amount of variability, 89.91% (FST; PCA1=57.89, PCA2=17.99 and PCA3=14.03), 87.63% (Dk; PCA1=60.92, PCA2=15.34 and PCA3=11.37) and 86.00% (RST; PCA1=46.45, PCA2=25.91 and PCA3=13.64), was accounted for in the first three PCA variables. All three plots (see Figure 2) showed a relatively similar pattern. The plot based on the FST distance matrix (Figure 2a) illustrated a separation of the 11 breeds into five geographic and genealogic groups, of which the ‘Noriker’ group and the ‘Rhenish’ group were the most distinctive; the ‘Croatian’ group was in the middle, while the Black Forest horse and Schleswig Draught horse populations formed two separate out-groups. In the plot based on molecular kinship distance Dk (Figure 2b), the pattern did not change much, except for the reorientation of both the mentioned out-groups. In the plot based on RST distance (Figure 2c), the Posavina breed has shifted towards the ‘Noriker’ group, and the close relationship between ‘Rhenish’ populations has stretched.
Three-dimensional plots illustrating genetic relationship among 11 sampled draught horse populations (434 horses), based on 27 microsatellite loci. PCA axes are the result of the principal component analysis (PCA) performed on different molecular distance matrices: (a) FST (Weir and Cockerham, 1984, b) Dk (Caballero and Toro, 2002) and (c) RST (Slatkin, 1995). Altmaerkisch Draught horse – A; Mecklenburg Coldblood – M; Saxon Thuringa Coldblood – ST; Rhenish German Draught horse – R; Black Forest horse – BF; Schleswig Draught horse – Sch; South German Coldblood – SG; Noriker-Carinthia – NC; Noriker-Salzburg – NS; Posavina horse – P and Croatian Coldblood – C.
Population structure and admixture level
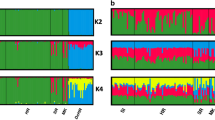
To measure the population structure and degree of admixture, we applied STRUCTURE, as the implemented algorithm uncovers ‘hidden structure’ without using a priori knowledge about the number of clusters (populations or breeds) present in a dataset. In order to illustrate a decision on the most likely number of clusters present in the dataset (the most likely parameter K), in Figure 3, we presented ln Pr(G∣K) values for all STRUCTURE runs. The most likely K is that where ln Pr(G∣K) is maximised. While all runs from K=5 to 9, as illustrated in Figure 4, showed a pattern that had a meaningful explanation, there was no clear ‘plateau’ (see Figure 3). The highest ln Pr(G∣K) value was obtained for K=9. On the other hand, the highest mean ln Pr(G∣K) value was obtained for K=8, as these values were more consistent than values obtained for K=9 (see Figure 3). Furthermore, the only difference between K=8 and 9 was that nine horses within the Rhenish German Draught horse formed a separate cluster in K=9 (see Figures 3 and 4). Assuming that those nine horses might have been of ‘problematic’ origin, we considered K=8 as being the number of clusters that capture the major structure of the data (representative K-value). Therefore, all further explanations are based on the STRUCTURE results obtained for K=8. The Black Forest horse, Rhenish German Draught horses, Schleswig Draught horse and South German Coldblood populations were identified as separate clusters (Figure 4), while the Altmaerkisch Coldblood, Mecklenburg Coldblood and Saxon Thuringa Coldblood populations were identified as one cluster. Noriker Carinthia and Noriker Salzburg populations were also clustered together (Figure 4). While the corresponding eight clusters fully matched one or several a priori classifications, the Croatian Coldblood and Posavina horse showed an admixture pattern between two distinct clusters (Figure 4).
Ln Pr(G∣K) values presented as a function of the number of clusters (Prichard et al., 2000; Falush et al., 2003). The largest ln Pr(G∣K) values within each K (among 10 runs) are presented with black circles.
Graphical presentations of the population structure analyses for a sample of 434 draught horses (a priory defined 11 populations). Each horse is represented by a single vertical line broken into K colour segments, with lengths proportional to the estimated membership of the inferred cluster. Among 10 runs, only graphical presentations for runs with the largest ln Pr(G∣K) values are shown.
The results of the analysis performed on the reduced datasets (‘Rhenish’ group, ‘Noriker’ group and ‘Croatian’ group) have shown the same pattern and confirmed the results presented.
Discussion
Population structure
Principal component analyses and graphic presentations showed that the analysed populations form five separate groups. The results obtained are concordant with historical evidence that includes the Altmaerkisch Coldblood, Mecklenburg Coldblood, Rhenish German Draught horses and Saxon Thuringa Coldblood in a single breed. It also considers the South German Coldblood to be a member of the ‘Noriker’ group (Aberle et al., 2004). On the other hand, the Black Forest horse was found to be quite distinctly separated from the ‘Noriker’ group. Aberle et al. (2004) showed that two populations, the South German Coldblood and the Black Forest horse, were genetically distinctive. Here, we have shown that Black Forest horse is also genetically quite distinctive from the Austrian Noriker horse. The closest breeds to the Black Forest horse were the Croatian Coldblood, South German Coldblood and Posavina horse with pairwise FST values: 0.063, 0.070 and 0.072, respectively. It seems that open herd management and introgression of other breeds has had a large influence on the breed. For example, Haflinger, Freiberger and Noriker horses occur in the pedigrees of the Black Forest horse. Two Croatian populations have been placed together, posing the question of their status (one-breed versus two-breed hypothesis). Until now, this topic has not been analysed by means of molecular markers. The Posavina horse is described as a well-adapted local, autochthonous breed, established by crossing Bosnian ponies and heavy horses with some influence of Lipizzan and Nonius blood. Croatian Coldblood is considered to be a local crossbreed of different draught horse breeds, in the beginning with Noriker horses and later with Ardennais, Belgian, Percheron and similar breeds. Croatian Coldblood is bred with an open herd book, while the Posavina horse herd book is closed. In a closed herd book, the parents must also be registered in this or another registry (acceptable by breeding organisation), while in an open herd book, animals can be registered without their parents having been previously registered.
The position of the Schleswig Draught horse shows the separation, which is concordant with written evidence as well as with geographical location. The results obtained from a posterior full Bayesian approach (STRUCTURE program) showed even better resolution, as they were able to detect further differentiations and provide answers to the specific conservation hypotheses. First, the ‘Rhenish’ group was separated into two clusters, a cluster with the Rhenish German Draught horse and a cluster with three other populations (Altmaerkisch Coldblood, Mecklenburg Coldblood and Saxon Thuringa Coldblood). The observed separation is a consequence of a genetic drift observable since the political separation of Germany in 1949. On the other hand, no clear differentiation could be determined among the other three ‘East German’ populations, recently separated into different breeding organisations. The obtained results are in agreement with those of Aberle et al. (2004), where, based on pairwise gene frequency comparisons, it has been shown that the Mecklenburg and Saxon Thuringa Coldblood are significantly different from the Rhenish German Draught horse, but not from each other.
Although shown to be closely related to the ‘Noriker’ group (pairwise FST values below 0.04, see Table 3), STRUCTURE was able to separate the South German Coldblood from the Austrian Noriker populations (see Figure 4 for K>6), but not the Noriker Carinthia from the Noriker Salzburg population (see Figure 4 for all K-values). While the Salzburg population is considered as an original breeding stock producing nearly all stallions used in Austria, up to the 1960s, the Carinthia population was considered as a different breed. The mentality of German breeders is characterised by the use of open herd books, whereas Austrian Noriker breeders claim to have had a closed herd book for nearly 300 years. Thus, the immigration of Thoroughbred and Warmblood into the South German Coldblood is a possible reason for this subtle differentiation within the ‘Noriker’ group. In the case of Croatian breeds, it was possible to detect the hidden admixture and to prove a mischaracterisation made by breeding organisations. The results presented show that the Croatian Coldblood and the Posavina horse are two separate, but closely related breeds (populations) that might be characterised and differentiated by means of molecular techniques (microsatellites) and advanced Bayesian methods (STRUCTURE algorithm). Overall, the results obtained by STRUCTURE indicate that political barriers (administrative formation of states) are an important factor influencing the differentiation among draught horse populations.
Conservation unit, genetic diversity and management strategies
When the history of a breed is complex, as in the case of draught horses involved in this study, it is difficult or even impossible to define breeds as clearly distinctive units. In such situations, the combined results of molecular and statistical approaches might provide a first insight into further conservation strategies that should be considered. In addition, those techniques might enable population (cluster) characterisation that has not been recognised previously. Thus, the pairwise FST differentiation of the populations reconstructed according to STRUCTURE estimated membership is shown in Table 3 (below the diagonal), while genetic diversity within hypothetical populations representing conservation units is shown in Table 4.
With respect to allelic richness, effective population size and their genetic closeness (see Figures 3 and 4), four populations of the ‘Rhenish’ group (Altmaerkisch Coldblood, Mecklenburg Coldblood, Rhenish German Draught horse and Saxon Thuringa Coldblood) might be considered as one breeding group consisting of four subpopulations. Technically, as demonstrated by STRUCTURE, it is possible to separate the Rhenish German Draught horse, but considering its low genetic diversity, the justification of such a procedure is questionable. Still, if such a decision were made, we would recommend planned migrations of horses from Altmaerkisch Coldblood, Mecklenburg Coldblood and Saxon Thuringa Coldblood populations to the Rhenish German Draught horse. The situation with the ‘Noriker’ group is somewhat different, because since those populations seem to be large enough, the merging of South German Coldblood with the Austrian Noriker horse (shown to be one unit) would not improve genetic diversity (see Table 4).
Furthermore, for ‘Croatian’ horses the STRUCTURE algorithm was able to recognise two clusters (FST=0.035; see Table 3), opening up new possibilities for the breeding of Croatian Coldblood and Posavina horses as two separate units according to their genetic purity. If this decision was taken, one should be aware that such a procedure would decrease the genetic diversity within these two populations (see Table 4). On the other hand, even when separated according to STRUCTURE membership index (q>0.70), the two populations were still closely related (FST=0.035). We propose merging those two populations into one conservation unit with two clearly defined subpopulations (Croatian Coldblood and Posavina horse) or separating those two populations as two conservation units (according to their genetic purity) and allowing substantial planned intermixing (long-term differentiation).
Two populations, the Black Forest horse and the Schleswig Draught horse, showed a relatively low allele richness and effective population size. Unfortunately, these two populations were quite distinctive from the other draught horse populations analysed in this study. Further genetic analyses would be required to find populations genetically more closely related to these two breeds. Considering historical evidence, it would be of further interest to analyse the relationship of the Black Forest horse with the Haflinger and Freiberger horses as well as the relationship of the Schleswig Draught horse with the Jutland and Suffolk Punch horses.
In order to take a more general and precise view on the conservation status of European draught horse populations, it would be necessary to extend this study to a large number of other breeds. Thus, for example, it would be of interest to include in the analysis Ardennais, Belgian Heavy Draught, Clydesdale, Friesian horse, Italian Heavy Draught, Irish Draught, Murgese, Norman Cob, Percheron, Russian Heavy Draught and Shire as well as other European working horse breeds. As, for thousands of years, draught horses have been an important force in changing the course of human civilization, and as they still have enormous potential, such an analysis would surely justify all the costs required.
Limitations
Given that conservation genetics is a complex issue, our intention was not to advocate application of specific conservation decisions. Moreover, we wanted to present facts that would aid in making efficient conservation decisions. In addition, we would like to stress that the presented results are based on the assumption that microsatellites represent neutral genetic variation. Thus, they do not relate to differences among and variability within populations with respect to agriculturally important traits, in most cases inherited according to quantitative genetic principals, which have undergone artificial or natural (adaptive) selection. Those traits might have additional value from a human or long-term adaptive perspective and should also be considered in future conservation strategies and programmes.
References
Aberle KS, Hamann H, Drögemüller C, Distl O (2004). Genetic diversity in German draught horse breeds compared with a group of primitive, riding and wild horses by means of microsatellite DNA markers. Anim Genet 35: 270–277.
Achmann R, Curik I, Dovc P, Kavar T, Bodo I, Habe F et al. (2004). Microsatellite diversity, population subdivision and geneflow in the Lipizzan horse. Anim Genet 35: 285–292.
Alvarez I, Gutierrez JP, Royo LJ, Fernandez I, Gomez E, Arranz JJ et al. (2005). Testing the usefulness of the molecular coancestry information to assess genetic relationships in livestock using a set of Spanish sheep breeds. J Anim Sci 83: 737–744.
Bailey E, Skow L, Bernoco D, DelValle A, Scavone MD, Bowling AT et al. (2000). Equine dinucleotide repeat loci LEX071 through LEX078. Anim Genet 31: 286–287.
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996–2004). GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier France.
Breen M, Lindgren G, Binns MM, Norman J, Irvin Z, Bell K et al. (1997). Genetical and physical assignments of equine microsatellites-first integration of anchored markers in horse genome mapping. Mammal Genome 8: 267–273.
Caballero A, Toro MA (2002). Analysis of genetic diversity for the management of conserved subdivided populations. Conserv Genet 3: 289–299.
Chikhi L, Bruford M (2005). Mammalian population genetics and genomics. In: Ruvinski A, Graves JM (eds). Mammalian Genomics, Chapter 21. CABI Publishing: Wallingford. pp 539–584.
Coogle L, Bailey E (1999). Equine dinucleotide repeat loci LEX064 through LEX070. Anim Genet 30: 71–72.
Coogle L, Bailey E, Reid R, Russ M (1996a). Equine dinucleotide repeat polymorphisms at loci LEX002, -003, -004, -005, -007, -008, -009, -010, -011, -013 and -014. Anim Genet 27: 126–127.
Coogle L, Bailey E, Reid R, Russ M (1996b). Equine dinucleotide repeat loci from LEX025 to LEX033. Anim Genet 27: 289–290.
Coogle L, Bailey E (1997). Equine dinucleotide repeat loci LFX049-LEX063. Anim Genet 28: 378.
Coogle L, Bailey E, Reid R, Russ M (1997). Equine dinucleotide repeat loci LEX034-LEX048. Anim Genet 28: 309.
Cothran EG, Luis C (2005). Genetic distance as a tool in the conservation of rare horse breeds, In: Bodo I, Alderson L, Langlois B (eds). Conservation Genetics of Endangered Horse Breeds. Wageningen Academic Publishers: Wageningen. EAAP Scientific Series no. 116, pp 55–72.
Ellegren H, Johansson M, Sandberg K, Andersson L (1992). Cloning of highly polymorphic microsatellites in the horse. Anim Genet 23: 133–142.
Glowatzski-Mullis ML, Muntwyler J, Pfister W, Marti E, Rieder S, Poncet PA et al. (2006). Genetic diversity among horse populations with a special focus on the Franches-Montagnes breed. Anim Genet 37: 1–7.
Godard S, Vaiman D, Oustry A, Nocart M, Bertaud M, Guzylack S et al. (1997). Characterization, genetic and physical mapping analysis of 36 horse plasmid and cosmid-derived microsatellites. Mammal Genome 8: 745–750.
Guerin G, Bertaud M, Amigues Y (1994). Characterization of seven new horse microsatellites: HMS1, HMS2, HMS3, HMS5, HMS6, HMS7 and HMS8. Anim Genet 25: 62.
Guo SW, Thompson EA (1992). Performing the exact test of Hardy–Weinberg proportions for multiple alleles. Biometrics 48: 361–372.
Gutiérrez JP, Royo LJ, lvarez I, Goyache F (2005). MolKin v2.0: a computer program for genetic analysis of populations using molecular coancestry information. J Hered 96: 718–721.
Hall SJG, Bradley DG (1995). Conserving livestock breed diversity. Trends Ecol Evol 10: 267–270.
Hardy OJ, Vekemans X (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Hill WG (1981). Estimation of effective population size from data on linkage disequilibrium. Genet Res 38: 209–216.
Hochberg Y (1988). A sharper Bonferroni procedure for multiple significance testing. Biometrica 75: 800–803.
Hopman TJ, Han EB, Story MR, Schug MD, Aquadro CF, Bowling AT et al. (1999). Equine dinucleotide repeat loci COR001-COR020. Anim Genet 30: 225–226.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Josche A (1878). Die Pferdezucht in Oesterreich-Ungarn nach ihren Haupt-Typen dargestellt. Klagenfurt: Austria.
Kakoi H, Tozaki T, Hirota K, Mashima S (1999). Genetic polymorphisms of equine microsatellite loci: TKY16, TKY19 and TKY21. Anim Genet 30: 68–69.
Meyer AH, Valberg SJ, Hillers KR, Schweitzer JK, Mickelson JR (1997). Sixteen new polymorphic equine microsatellites. Anim Genet 28: 69–70.
Murphie AM, Hopman TJ, Schug MD, Aquadro CF, Bowling AT, Murray JD et al. (1999). Equine dinucleotide repeat loci COR021-COR040. Anim Genet 30: 235–237.
Miraglia N, Burger D, Kapron M, Flanagan J, Langlois B, Martin-Rosset W (2006). Local animal resources and products in sustainable development: role and potential of equids. In: Rubino R, Sepe L, Dimitriadou A, Gibon A (eds). Livestock Farming Systems – Product Quality Based on Local Resources Leading to Improved Sustainability. Wageningen Academic Publishers. Wageningen. EAAP Scientific Series no. 118, pp 217–234.
Pearse D, Crandall KA (2004). Beyond FST: Analysis of population genetic data for conservation. Conserv Genet 5: 585–602.
Peel D, Ovenden JR, Peel SL (2004). NEESTIMATOR: software for estimating effective population size, Version 1.3. Queensland Government, Department of Primary Industries and Fisheries, St Lucia, Queensland.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Rosenberg NA (2004). DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138.
Ruth LS, Hopman TJ, Schug MD, Aquadro CF, Bowling AT, Murray JD et al. (1999). Equine dinucleotide repeat loci COR041-COR060. Anim Genet 30: 320–321.
SAS Institute Inc. (1999–2001). SAS/STAT Software: Changes and Enhancements Through Release 8.02. Cary, NC, USA.
Slatkin M (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462.
Swinburne JE, Lockhart L, Aldridge V, Marti E, Breen M, Binns MM (2000). Characterisation of 25 new physically mapped horse microsatellite loci: AHT24-48. Anim Genet 31: 237–238.
Tallmadge RL, Hopman TJ, Schug MD, Aquadro CF, Bowling AT, Murray JD et al. (1999). Equine dinucleotide repeat loci COR061-COR080. Anim Genet 30: 462–463.
Van Haeringen H, Bowling AT, Stott ML, Lenstra JA, Zwaagstra KA (1994). A highly polymorphic horse microsatellite locus: VHL20. Anim Genet 25: 207.
Van Haeringen WA, van de Goor LH, van der Hout N, Lenstra JA (1998). Characterization of 24 equine microsatellite loci. Anim Genet 29: 153–156.
Weir BS, Cockerham CC (1984). Estimating F-Statistics for the analysis of population structure. Evolution 38: 1358–1370.
Acknowledgements
We are grateful to everyone who helped with data collection (Gertrud Grilz-Seger, Kresimir Salajpal and Mato Cacic). We also thank Roland Achmann, Felix Goyache, Juan Pablo Gutierrez and two anonymous reviewers for the valuable comments and suggestions that improved the manuscript. Finally, we would like to thank Dragan Tupajic for the graphic design of the figures. This work was financially supported by Austrian Ministry of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Druml, T., Curik, I., Baumung, R. et al. Individual-based assessment of population structure and admixture in Austrian, Croatian and German draught horses. Heredity 98, 114–122 (2007). https://doi.org/10.1038/sj.hdy.6800910
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800910
Keywords
This article is cited by
-
Genetic diversity of a large set of horse breeds raised in France assessed by microsatellite polymorphism
Genetics Selection Evolution (2009)