Abstract
The Viking age witnessed the expansion of Scandinavian invaders across much of northwestern Europe. While Scandinavian settlements had an enduring cultural impact on North Atlantic populations, the nature and extent of their genetic legacy in places such as Shetland and Orkney is not clear. In order to explore this question further, we have made an extensive survey of both Y-chromosomal and mitochondrial DNA (mtDNA) variation in the North Atlantic region. Our findings indicate an overall Scandinavian ancestry of ≈44% for Shetland and ≈30% for Orkney, with approximately equal contributions from Scandinavian male and female subjects in both cases. This contrasts with the situation for the Western Isles, where the overall Scandinavian ancestry is less (≈15%) and where there is a disproportionately high contribution from Scandinavian males. In line with previous studies, we find that Iceland exhibits both the greatest overall amount of Scandinavian ancestry (55%) and the greatest discrepancy between Scandinavian male and female components. Our results suggest that while areas close to Scandinavia, such as Orkney and Shetland, may have been settled primarily by Scandinavian family groups, lone Scandinavian males, who later established families with female subjects from the British Isles, may have been prominent in areas more distant from their homeland.
Similar content being viewed by others
Introduction
Abundant evidence from archaeology, oral and written histories, and place-names bears witness to an extensive Scandinavian cultural impact on the island populations of the North Atlantic (Jones, 1984; Graham-Campbell and Batey, 1998; Sawyer, 1999). The scale and the nature of the Scandinavian genetic legacy in this region, however, are less well understood. Viking exploits in the British Isles are thought to have begun with the sacking of the monastery at Lindisfarne off the Northumbrian coast as early as AD 793, followed by attacks on Jarrow (794) and the Columban church of Iona (976, 802, 806). Scandinavian Vikings soon dominated the sea routes and coastlines stretching from Norway to Shetland, Orkney, Scotland, the Hebrides and Ireland. The period of Scandinavian political and cultural domination in this region lasted until 1472 when Orkney and Shetland became part of Scotland.
Early attempts to estimate the genetic impact of Scandinavian settlement on local populations of the North Atlantic region were mostly based on analyses of serogenetic variants from contemporary populations, which gave conflicting or unclear results (Thompson, 1973; Morton et al, 1977; Wijsman, 1984). One problem with using such markers is that they provide little power to discern identity by descent – a crucial issue in studies of genetic relationships among individuals. In recent years, haploid, nonrecombining and uniparentally inherited genetic systems from the Y-chromosome and mitochondrial genome have become standard tools for genetic analyses of human populations (Passarino et al, 2002; Gonzalez et al, 2003; Jobling and Tyler-Smith, 2003). Although patrilineal (Y-chromosome) and matrilineal (mtDNA) genealogical pathways only represent a fraction of all genealogical pathways between a given set of individuals, these genealogies are entirely nonoverlapping and discrepancies between them thus have a clear interpretation, that is differences in the genetic history of the sexes.
Recent studies of Y-chromosomes and mtDNA of Icelanders indicate that this population's gene pool was the result of admixture between male subjects whose patrilineal ancestry lay in Scandinavia, and female subjects whose matrilineal ancestry lay within the British Isles (Helgason et al, 2000a, 2000b, 2001). A further study suggests that Icelanders and Orkney Islanders have similar proportions of Scandinavian mtDNA ancestry (≈36%; Helgason et al, 2001). Other studies of the genetic history of the North Atlantic region have concentrated on discerning the different proportions of northwestern European, Anglo-Saxon and Scandinavian patrilineal ancestry using Y-chromosome data (Wilson et al, 2001; Weale et al, 2002; Capelli et al, 2003). A sizeable component of Scandinavian patrilineal ancestry has been reported in Orkney (55%) and Shetland (68%) based on likelihood estimates of population admixture and principal components analyses of haplotype frequencies (Capelli et al, 2003).
Here, we present an analysis of new mtDNA and Y-chromosome data from Shetland and new Y-chromosome data from Scotland, Norway, the Western Isles and the Isle of Skye. We combine our new data with those that have been previously published in order to investigate further the relationships among populations of the North Atlantic, and to establish what genetic legacy remains from Scandinavian activities in this area during the Viking period. In particular, our aim is to evaluate the different genetic impact of Scandinavian male and female subjects on Shetland and other island and coastal populations in the North Atlantic region.
Methods
DNA extractions were made from 503 new buccal swab samples collected by the authors in Shetland using specifically designed Biorad™ kits. The hypervariable segment 1 (HVS1) of the mtDNA control region was sequenced between sites 16055 and 16383 for each of these samples, according to previously described methods (Richards et al, 1996) and submitted to GenBank (accession numbers AY950803–AY951304). A total of 192 Shetland males from the above sample were typed for five Y-chromosome biallelic loci (Rosser et al, 2000; 92R7, M9, SRY1532, TAT and YAP) and five Y-chromosome microsatellites (Kayser et al, 1997; DYS19, DYS390, DYS391, DYS392 and DYS393). In addition, previously collected DNA samples from the Scottish mainland (366), Scottish northwestern coastal region (103) and the Western Isles and Isle of Skye (72) were analyzed for the same 10 Y-chromosome loci. These 733 new Y-chromosome haplotypes (which are available as an online supplement) include 61 that have been previously described (Helgason et al, 2000a).
In addition to samples typed by the authors for this study, the following previously published mtDNA HVS1 sequences were used: 398 Icelanders (Helgason et al, 2000b), 551 Icelanders (Helgason et al, 2003), 39 Icelanders (Sajantila et al, 1995), 78 Orkney Islanders (Helgason et al, 2001), 664 inland Scots (Helgason et al, 2001), 216 northwestern coastal Scots (Helgason et al, 2001), 226 inhabitants from the Western Isles and the Isle of Skye (Helgason et al, 2001), 105 Irish (Richards et al, 2000), 216 Norwegians (Opdal et al, 1998), 324 Norwegians (Helgason et al, 2001), 74 Norwegians (Passarino et al, 2002), 28 Swedes (Kittles et al, 1999) and 32 Swedes (Sajantila et al, 1996). The following previously published Y-chromosome haplotypes were also used: 181 Icelanders (Helgason et al, 2000a), 12 Danes (Helgason et al, 2000a), 221 Irish (Helgason et al, 2000a), one Irish (Hurles et al, 1999), 110 Norwegians (Helgason et al, 2000a), two Norwegians (Hurles et al, 1999), 78 Norwegians (Wilson et al, 2001), 71 Orkney Islanders (Wilson et al, 2001) and 109 Swedes (Zerjal et al, 2001).
The Scandinavian source population is represented in our study by contemporary samples from Norway, Sweden and Denmark and the source population of the Northern and Western British Isles is represented by contemporary samples from Ireland and inland Scotland. We examine the contribution of these two source populations to five putative admixed island and coastal populations: Shetland, Orkney, Western Isles/Isle of Skye, Iceland and the northwestern coastal region of Scotland (defined as the coastal zones from the Moray Firth to Kintyre).
For analyses based on the mutational differences between Y-chromosome haplotypes, mutations at biallelic loci were weighted by a factor of 100 relative to microsatellite loci. Analysis of molecular variance (AMOVA) calculations and exact tests of population differentiation were performed using the Arlequin v. 2.0 software package (Schneider et al, 2000). Gene diversity measures were made using the following standard formula: (n/[n−1])(1−∑i=1kpi2), where n represents sample size, k the number of distinct haplotypes and pi the frequency of haplotype i. The population-mutation-rate parameter, θk, was estimated using Ewens' sampling formula (Ewens, 1972). This parameter assumes selective neutrality of the genetic markers, an infinite alleles mutation model and constant population size. Admixture was estimated using the mρ method (Helgason et al, 2001). The best fitting admixture model is found by searching over the line 0≤ηN≤1 of possible admixture models, where ηN represents the proportion of Scandinavian founders. The probability that a randomly chosen haplotype from the admixed population originated from the Scandinavian source population is determined by its relative frequency in the two source populations (pN and pG being the respective Scandinavian and British/Irish frequencies of the haplotype) with the model being tested as follows: ηNpN[ηNpN+(1−ηN)pG]. If the chosen haplotype is not present in the source populations, this probability is derived from the relative frequency of the closest matching haplotypes in terms of mutational differences (the presumed founder haplotypes). For each admixture model, 5000 runs of a Monte Carlo simulation were performed, where each haplotype from the admixed population was assigned an origin on the basis of the above-described probability, yielding an average proportion of haplotypes assigned a Scandinavian origin (mρN). The fit of the model to the outcome of the simulations was evaluated by

where N is the number of haplotypes sampled from the admixed population.
Results
Population structure
The extent and nature of population structure in mtDNA and Y-chromosome variation within the North Atlantic region was evaluated using AMOVA statistics based on the mutational differences between haplotypes (Excoffier et al, 1992) and an exact test of population differentiation based on haplotype frequencies (Raymond and Rousset, 1995). Out of the total genetic variance exhibited by the 3454 mtDNA sequences from the North Atlantic region, only 0.66% was attributable to differences among the seven populations. This small FST value (0.0066) was nonetheless shown to be significantly different from 0 (P=0) by means of randomization of haplotypes among populations. This suggests that although the degree of phylogeographic structuring of mtDNA variation identified is small, the distribution of haplotypes among populations nevertheless deviates from random expectation. This conclusion is supported by an exact test of sample differentiation (using 10 000 Markov steps), which shows a significant difference (P=0) between the haplotype distributions of all pairs of populations, with the following exceptions: (1) Orkney vs Norway and Sweden, (2) Orkney vs Scotland and Ireland, (3) Orkney vs Scottish N&W coastline and (4) Scottish N&W coastline vs Scotland and Ireland. We note that the relatively small sample of mtDNA sequences from Orkney may partially account for the apparent lack of differentiation from other populations.
AMOVA analyses suggest a greater level of differentiation among populations for the 1521 Y-chromosomes sampled in this study, than was detected for mtDNA. For the Y-chromosomes, 8.12% of the total genetic variance was attributable to differences among the seven populations, with the FST value (0.0812) shown to be significantly different from 0 by means of randomization as before (P=0). All pairs of populations exhibited statistically significant differences (P<0.05) in the distribution of Y-chromosome haplotypes according to exact tests, with the exception of (1) Western Isles/Skye vs Scotland and Ireland, and (2) Western Isles/Skye vs Scottish N&W coastline.
The results of the exact tests (which do not exploit mutational differences among types) suggest that the population structure that is detected in both mtDNA and Y-chromosome variation is due to the differential distribution of individual haplotypes (located at the tips of shared, deep-rooted phylogenetic clades), rather than to differences in the differential distribution of deep-rooted clades among populations.
Genetic diversity
Measures of mtDNA and Y-chromosome variation in the populations of the North Atlantic region are presented in Tables 1 and 2. It is important to note that gene diversity and other pairwise measures of genetic diversity are unreliable indicators of varying effective population size among closely related, recently diverged populations such as those in this study (Helgason et al, 2003). In contrast, statistics based on the number of haplotypes (θk) and the average number of haplotypes obtained through iterative resampling have been shown to be more reliable indicators of differences in effective population size (Helgason et al, 2003). For the current data sets, these latter statistics present a picture of Y-chromosome and mtDNA diversity that is largely consistent with expectations based on historical and archaeological evidence relating to population size in the North Atlantic region. Thus, smaller, geographically isolated island populations have fewer haplotypes per number of individuals sampled than mainland populations, which are likely to have a larger effective population size.
Differences in the proportions of private haplotypes can be informative about the relative importance of (1) effective population size and mutation rate (which combine to give the population mutation-rate parameter θ) and (2) gene flow (m), to the production and maintenance of genetic diversity. The proportion of private haplotypes encountered in a population is expected to be positively related to θ and negatively related to m (Helgason et al, 2001). Given that we have an independent estimate of θ based on the number of haplotypes encountered per individuals sampled (θk as defined by Ewens, 1972), we can regress the proportion of private haplotypes against θk for a set of populations. Deviations from the resulting regression line are related to the amount of gene flow that each population has received, although the relationship can be distorted both by sampling error and violations of assumptions implicit in estimates of θk (ie selective neutrality and constant population size).
For mtDNA, the correlation between θk and the proportion of private lineages is r=0.77 (P=0.041), with only two populations found outside the 95% confidence region around the regression line. Iceland has a relative excess of private haplotypes, which is indicative of genetic isolation and consistent with the fact that this is the most geographically remote population in the analysis. In contrast, the inhabitants of the Scottish N&W coastline have a relative deficit of private mtDNA haplotypes. This suggests a higher level of female-mediated gene flow to and from these islands and is consistent with the fact that they are the least geographically isolated population in the current study. For Y-chromosomes, the correlation between θk and the proportion of private lineages is stronger than for mtDNA, r=0.925 (P=0.003). Only one population, the Western Isles/Isle of Skye, lies outside the 95% confidence region around the regression line, indicating a deficit of private Y-chromosome haplotypes. This deficit can be interpreted in terms of the homogenizing effect of male-mediated gene flow between these islands and the mainland.
Scandinavian and British/Irish ancestry in the island and coastal populations of the North Atlantic region
The analysis of population structure presented above indicates significant differences between the Y-chromosome and mtDNA pools of the two putative parental populations of Britain/Ireland and Scandinavia (represented by the contemporary inhabitants of Norway, Sweden and Denmark, and Scotland and Ireland, respectively). As a consequence, it should be possible to distinguish between the contributions that each of the two parental populations has made to the Y-chromosome and mtDNA gene pools of the island and coastal populations of the North Atlantic region. Estimates of population admixture can be made using one of several different approaches; as a consequence, the choice of statistical method used here necessitates some comment.
Standard frequency-based methods of admixture estimation assume that all alleles present in the hybrid population are also present in at least one of the putative parental populations (Roberts and Hiorns, 1965; Long, 1991). Such methods are unlikely to be informative for the current molecular data sets because as many as half of the Y-chromosome and mtDNA haplotypes in the hybrid populations are not found in either of the parental populations. An alternative method, which is based on average mutational differences between haplotypes drawn within or between admixed and parental populations, does not require any of the haplotypes from the hybrid population to be present in the parental populations. However, this method is only effective in cases where there is a marked mutational divergence between haplotypes from the parental populations (Bertorelle and Excoffier, 1998). This approach is not appropriate for the current study because the phylogenetic divergence of haplotypes from the parental populations is negligible, particularly for mtDNA.
An alternative method, mρ, was used in order to circumvent the problems inherent in the methods described above. mρ is a heuristic, frequency-based admixture approach that uses a distance matrix between haplotypes to identify founder haplotypes in the parental populations for all private haplotypes in hybrid populations. This method also takes into account uncertainty in estimates of haplotype frequencies in the hybrid and parental populations due to either sampling variance or genetic drift subsequent to population mixing (Helgason et al, 2001).
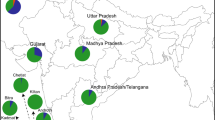
Table 3 and Figure 1 show estimates of Scandinavian and British/Irish ancestry based on the mρ approach. The proportion of Scandinavian patrilineal (Y-chromosome) ancestry is highest in Iceland (75%), is somewhat lower in Shetland (44.5%) and Orkney (31%), declines further in the Western Isles and Skye (22.5%) and is lowest along the N&W coastline of Scotland (15%). This is consistent with a lasting genetic legacy of Scandinavian males in both Orkney and Shetland with a lesser, but still measurable, contribution to the other Scottish locations.
Map showing the proportions of Scandinavian and British/Irish ancestry for mtDNA (Mt) and Y-chromosomes (Y) for each of the admixed populations from the North Atlantic region included in this study. The Scottish island groups of Shetland, Orkney, the Western Isles and Skye, and the region that we define as the ‘North and West coast of Scotland’, are encircled for clarity.
The proportions of Scandinavian matrilineal (mtDNA) ancestry estimated for Shetland, Orkney and the N&W coastline of Scotland are almost identical to their patrilineal counterparts. In contrast, the estimated contribution of Scandinavian female subjects to the Western Isles and Isle of Skye (11%) is half that of Scandinavian male subjects (∼22%). This discrepancy is even more marked for Icelanders, whose effective founding population appears to have consisted largely of female subjects with a British/Irish ancestry and male subjects with a Scandinavian ancestry (Helgason et al, 2000a, 2001).
Our estimates of Scandinavian patrilineal ancestry for Shetland, Orkney and the Western Isles are smaller than those proposed by Capelli et al (2003), which were based on a different data set (their estimates were 68.3, 55.3 and 61.6%, respectively). We note, however, that while they employed a frequency-based admixture method that accounted for sampling variance and the impact of genetic drift, this method ignored all private haplotypes in the hybrid populations. It is likely that the difference between our admixture results and those of Capelli et al (2003) stems in part from this omission of private haplotypes. However, the difference could to some extent also be due to the use by Capelli et al (2003) of Y-chromosomes from both central Ireland and the Basque region to represent the ‘Celtic’ parental population.
Discussion
As is common in cases where genetic data are used to make inferences about historical events, the admixture results obtained in the present study may be open to a number of lines of interpretation. There are, however, some unambiguous conclusions that can be drawn from the study and a number of likely historical explanations that may be proposed.
Our mitochondrial and Y-chromosome data suggest that Shetland has almost identical proportions of Scandinavian matrilineal and patrilineal ancestry (∼44%). The same balance is to be found in Orkney and the N&W coastline of Scotland, although the average Scandinavian contribution in these two areas is lower (∼30% in Orkney and ∼15% in the N&W Scottish coastline). The results strongly suggest that the Scandinavian genetic contribution (in terms of both matrilineal and patrilineal ancestry) is greatest in those areas that lie close to Scandinavia. While it is possible that genetic drift has affected gene frequencies so that our estimates of admixture no longer accurately reflect the composition of founder populations, it seems unlikely to be coincidental that the overall proportion of Scandinavian ancestry for these three locations decreases in harmony with geographic distance from Scandinavia. Admixture analysis (using mρ) of recently published Y-chromosome data from the Faroe Islands (Jorgensen et al, 2004) also provides evidence for a trend of increased Scandinavian patrilineal ancestry in regions further from the British Isles. The estimated Scandinavian genetic component of the Faroese samples is 87% (N=89, k=26), a higher figure than for either Shetland or Orkney. Furthermore, as expected for a region with a historical population size even smaller than that of the Icelanders and with a similar degree of geographical isolation, our estimates suggest that the Faroese have the least diverse Y-chromosome pool (gene diversity=0.9298, θk=11.99).
Historical, archaeological, place-name and linguistic evidence indicates complete Norse cultural dominance of Shetland and Orkney during the Viking period (Jones, 1984). Although more distant from the Scandinavian power-base, Orkney was the nucleus of Scandinavian political and cultural influence in the North Atlantic colonies (Davies, 1999). This implies a Scandinavian presence in Orkney that was at least as great as that in Shetland. If this is the case and the indigenous pre-Scandinavian populations of Shetland and Orkney were about the same size, then the lesser Scandinavian genetic component in Orkney may be explained by greater post-Scandinavian migration from Scotland to Orkney than to Shetland. However, a smaller indigenous pre-Scandinavian population in Shetland, followed by similar numbers of Scandinavian colonizers and post-Scandinavian Scottish settlers would also account for the difference between the two islands. The genetic evidence does not allow us to determine whether the observed geographic gradient of Scandinavian ancestry is largely due to either variable pre-Scandinavian population sizes, variable numbers of Scandinavian settlers, variable levels of post-Scandinavian Scottish resettlement or some combination of all of these factors. However, the observed symmetry between Scandinavian mtDNA and Y-chromosome ancestry in Shetland, Orkney and the N&W coastline of Scotland suggests that roughly equal numbers of Scandinavian male and female subjects were involved in the colonization of these locations. In other words, the genetic evidence indicates a family-based Scandinavian settlement of Shetland, Orkney and the N&W coastline of Scotland. This contrasts with results obtained for the Western Isles/Isle of Skye and Iceland where estimated male and female Scandinavian components differ. The Western Isles/Isle of Skye have the lowest estimated proportion of Scandinavian ancestry (17%) while Iceland has the highest (55%), but both exhibit a two-fold excess of Scandinavian patrilineal ancestry relative to Scandinavian matrilineal ancestry. The data seem to suggest that the settlement pattern of both the Western Isles/Isle of Skye and Iceland included, in addition to Scandinavian family groups, a substantial number of males who took wives from indigenous British populations.
Islands off the coast of Britain are relatively distant from the Scandinavian homeland and must have been viewed as frontier areas within the North Atlantic region during the Viking period. Iceland, an uninhabited island at the margin of the then known world, was even more remote. Linguistic, archaeological and historical evidence indicates that Scandinavian dominance was never complete or secure in places such as the British Isles (Davies, 1999). Viewed in this light, the excess of Scandinavian patrilineal ancestry observed in the Western Isles and Iceland and the symmetric contribution of Scandinavian male and female subjects in Shetland, Orkney and the N&W coastline of Scotland are consistent with a common feature of human colonizing behavior: This is that migration to insecure frontier areas tends to involve a disproportionate number of lone male colonizers, whereas family groups are more likely to be abundant in secure areas that are closer to the strongholds of colonial power.
Accession codes
References
Bertorelle G, Excoffier L (1998). Inferring admixture proportions from molecular data. Mol Biol Evol 15: 1298–1311.
Capelli C, Redhead N, Abernethy JK, Gratrix F, Wilson JF, Moen T et al (2003). A Y-chromosome census of the British Isles. Curr Biol 13: 979–984.
Davies N (1999). The Isles: A History. Macmillan: London.
Ewens WJ (1972). The sampling theory of selectively neutral alleles. Theor Popul Biol 3: 87–112.
Excoffier L, Smouse P, Quattro J (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Gonzalez AM, Brehm A, Perez JA, Maca-Meyer N, Flores C, Cabrera VM (2003). Mitochondrial DNA affinities at the Atlantic Fringe of Europe. Am J Phys Anthropol 120: 391–404.
Graham-Campbell J, Batey CE (1998). Vikings in Scotland: An Archaeological Survey. Edinburgh University Press: Edinburgh.
Helgason A, Hickey E, Goodacre S, Bosnes V, Stefánsson K, Ward R, Sykes B (2001). mtDNA and the Islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry. Am J Hum Genet 68: 723–737.
Helgason A, Nicholson G, Stefánsson K, Donnelly P (2003). A reassessment of genetic diversity in Icelanders: strong evidence from multiple loci for relative homogeneity caused by genetic drift. Ann Hum Gen 67: 281–297.
Helgason A, Sigur∂́adóttir A, Gulcher J, Ward R, Stefánsson K (2000b). MtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet 66: 999–1016.
Helgason A, Sigur∂́adóttir A, Nicholson J, Sykes B, Hill EW, Bradley DG et al (2000a). Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 67: 697–717.
Hurles ME, Veitia R, Arroyo E, Armenteros M, Bertranpetit J, Perez-Lezaun A et al (1999). Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet 65: 1437–1448.
Jobling MA, Tyler-Smith C (2003). The human Y-chromosome: an evolutionary marker comes of age. Nat Rev Genet 4: 598–612.
Jones G (1984). A History of the Vikings. Oxford University Press: Oxford.
Jorgensen TH, Buttenschön HN, Wang AG, Als TD, Børglum AD, Ewald H (2004). The origin of the isolated population of the Faroe Islands investigated using Y chromosomal markers. Hum Genet 115: 19–28.
Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G et al (1997). Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110: 125–133.
Kittles RA, Bergen AW, Urbanek M, Virkkunen M, Linnoila M, Goldman D et al (1999). Autosomal, mitochondrial, and Y chromosome DNA variation in Finland: evidence for a male-specific bottleneck. Am J Phys Anthropol 108: 381–399.
Long JC (1991). The genetic structure of admixed populations. Genetics 127: 417–428.
Morton NE, Dick HM, Allan NC, Izatt MM, Hill R, Yee S (1977). Bioassay of kinship in Northwestern Europe. Ann Hum Genet 41: 249–255.
Opdal SH, Rognum TO, Vege A, Stave AK, Dupuy BM, Egeland T (1998). Increased number of substitutions in the D-loop of mitochondrial DNA in the sudden infant death syndrome. Acta Paediatr 87: 1039–1044.
Passarino G, Cavalleri GL, Lin AA, Cavalli-Sforza LL, Borresen-Dale AL, Underhill PA (2002). Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms. Eur J Hum Genet 10: 521–529.
Raymond M, Rousset F (1995). An exact test for population differentiation. Evolution 49: 1280–1283.
Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A et al (1996). Paleolithic and Neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 59: 185–203.
Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V et al (2000). Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67: 1251–1276.
Roberts DF, Hiorns RW (1965). Methods of analysis of the genetic composition of a hybrid population. Hum Biol 37: 38–43.
Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A et al (2000). Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67: 1526–1543.
Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML et al (1995). Genes and languages in Europe: An analysis of mitochondrial lineages. Genome Res 5: 42–52.
Sajantila A, Salem AH, Savolainen P, Bauer K, Gierig C, Paabo S (1996). Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc Natl Acad Sci USA 93: 12035–12039.
Sawyer P (1999) In: Sawyer P (ed) The Oxford Illustrated History of the Vikings. Oxford University Press: Oxford. pp 1–18.
Schneider S, Roessli D, Excoffier L (2000). Arlequin: a software for population genetic data. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
Thompson EA (1973). The Icelandic admixture problem. Ann Hum Genet 37: 69–80.
Weale ME, Weiss DA, Jager RF, Bradmnam N, Thomas M (2002). Y chromosome evidence for Anglo-Saxon mass migration. Mol Biol Evol 19: 1008–1021.
Wijsman EM (1984). Techniques for estimating genetic admixture and applications to the problem of the origin of the Icelanders and the Ashkenazi Jews. Hum Genet 67: 441–448.
Wilson JF, Weiss DA, Richards M, Thomas M, Bradman N, Goldstein DB (2001). Genetic evidence for different male and female roles during cultural transitions in the British Isles. Proc Natl Acad Sci USA 98: 5078–5083.
Zerjal T, Beckman L, Beckman G, Mikelsaar AV, Krumina A, Kucinskas V et al (2001). Geographical, linguistic, and cultural influences on genetic diversity: Y-chromosomal distribution in Northern European populations. Mol Biol Evol 18: 1077–1087.
Acknowledgements
We thank all the volunteers who contributed their DNA to this study, the Scottish Blood Service for their help and both the Wellcome Trust and Medical Research Council for support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Heredity web site (http://www.nature.com/hdy)
Supplementary information
Rights and permissions
About this article
Cite this article
Goodacre, S., Helgason, A., Nicholson, J. et al. Genetic evidence for a family-based Scandinavian settlement of Shetland and Orkney during the Viking periods. Heredity 95, 129–135 (2005). https://doi.org/10.1038/sj.hdy.6800661
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800661
Keywords
This article is cited by
-
Subdividing Y-chromosome haplogroup R1a1 reveals Norse Viking dispersal lineages in Britain
European Journal of Human Genetics (2021)
-
Deeper Histories of Dispossession: The Genealogy of “Proletarian” Relations in Iceland
Historical Archaeology (2019)
-
The fine-scale genetic structure of the British population
Nature (2015)
-
Mitochondrial diversity patterns and the Magdalenian resettlement of Europe: new insights from the edge of the Franco-Cantabrian refuge
Journal of Human Genetics (2012)
-
Fine-mapping and candidate gene investigation within the PARK10 locus
European Journal of Human Genetics (2009)