Abstract
Rainforests in Australia and around the world have been extensively cleared and degraded. It is essential to recognize the changes in population diversity and dynamics that follow habitat fragmentation if better conservation and management strategies are to be developed. This study is an investigation of the medium term (over 100 years) effects of rainforest fragmentation on a long-lived, early successional tree species within a habitat matrix that includes various types of fragmented and undisturbed sites. Five microsatellite loci were used to assess the level and distribution of genetic variation across the southern range of Elaeocarpus grandis (Elaeocarpaceae). In all, 21 sites were sampled to provide a direct comparison between fragmented and undisturbed populations. Overall levels of diversity (A=3.4, He=0.568, f=0.094) were higher than those of closely related endemic species, but lower than those recorded across other rainforest trees. No significant genetic structure was detected across this species, suggesting the existence of efficient dispersal and colonization mechanisms responsible for the maintenance of gene flow. Rainforest fragments, and in particular those within the extensively cleared Big Scrub, show a trend for increased inbreeding levels caused by a loss of heterozygosity within juvenile cohorts. However, the overall rate of genetic decline within fragmented rainforests appears to be more subtle in E. grandis than across other species. A combination of ecological attributes and evolutionary history is likely to have contributed to this outcome and need to be considered in future rainforest restoration projects.
Similar content being viewed by others
Introduction
Rainforests worldwide are subject to increasing amounts of external pressure and degradation that dissect continuous species distributions into small, isolated fragments (Whitmore, 1997). Drastic environmental modifications threaten the integrity and long-term sustainability of ecosystems, as well as the survival of single species. As a result, research on the effects of habitat fragmentation has become a priority of the international conservation agenda, particularly within highly diverse ecosystems. An increasing body of knowledge on the changes in species composition across rainforest fragments, the effects of environmental attributes associated with disturbance, as well as natural and artificial re-colonization processes, is available (Laurence and Bierregaard, 1997), yet little is understood about the genetic requirements essential to the long-term survival of rainforest remnants.
After dominating most of the Australian continent, rainforests substantially declined from the mid-Miocene following major local and global climatic changes. During the Pleistocene glaciations, the remaining areas of rainforest were further subjected to contraction and expansion cycles influenced, among other things, by changes in fire frequencies (Bowman, 2000). To date, this vegetation type is mainly restricted to the eastern coast of the continent, but despite representing less than 1% of Australia's total vegetation cover it contains a considerable proportion of its biodiversity, including over 550 plant genera (Webb and Tracey, 1981). The rainforests of north eastern New South Wales (NSW) are included within the McPherson-Maclay Overlap, a very important biodiversity region that includes representatives of both tropical and temperate floras (Burbidge, 1960). Over 50 endemic genera are found in this region and a further 200+ species are at their most southern or northern limits (McDonald and Elsol, 1984).
Logging, clearing and urbanization in the last 150 years caused swift environmental degradation, particularly in lowland rainforests (Connelly and Specht, 1988). For instance, the ‘Big Scrub’ of northern NSW was the largest known area of continuous lowland subtropical rainforest in Australia, covering over 70 000 ha prior to European settlement (Lott and Duggin, 1993). Extensive clearing in the region since the mid-1800s has now reduced rainforest cover in the Big Scrub to approximately 1% of its original extent (Lott and Duggin, 1993). What remains forms a matrix of fragments varying in size, isolation and species diversity (sometimes still surprisingly high) that may not be evolutionary viable unless appropriate management actions are taken. This fragmented matrix creates an ideal experimental framework for examining the changes in population dynamics caused by habitat fragmentation, as it is easily accessible and surrounded by protected ‘control’ areas.
Biodiversity conservation within habitat fragments depends on the capacity of single species to survive bottlenecks and stochastic events. Conservation genetics can play an important role in interpreting the mechanisms threatening population survival within fragmented ecosystems (Templeton et al, 1990; Young and Clarke, 2000). As habitat fragmentation often reduces population size and increases isolation, shortfalls in genetic diversity and gene flow are to be expected. In fact, recent evidence suggests that reversing fragmentation processes through habitat management can have measurable effects on animal populations (Hale et al, 2001). A number of genetic processes, including inbreeding and drift, can ultimately reduce fitness and adaptation potential across the entire metapopulation, but while it can be assumed that genetic changes will occur the nature of those changes cannot always be predicted by simple theory (Young et al, 1996). Ultimately, only empirical investigations across representative taxa and within suitable study systems can demonstrate the changes in population dynamics caused by the artificial dissection of natural ecosystems.
The indispensable information on genetic variability, inbreeding rates and gene flow between and within populations can be efficiently gathered using highly informative molecular tools such as microsatellites (or simple sequence repeats – SSRs). These highly polymorphic and co-dominantly inherited markers are sensitive indicators of subtle changes in genetic diversity and inbreeding characteristics within small, isolated habitat fragments, and are therefore becoming the molecular tool of choice in population, fragmentation and ecological research. Frequent advances in experimental design and analytical approaches enable researchers to investigate gene exchange and population structuring at increasingly fine scales (eg Smouse and Peakall, 1999).
Elaeocarpus grandis is distributed throughout subtropical and tropical rainforests in Australia, often colonizing suitable disturbed areas but also persisting within the mature forest canopy. Owing to its fast growth rate, E. grandis is a good candidate for rainforest regeneration projects and future forestry ventures. It is estimated that prior to clearing, an ample habitat suitable to E. grandis existed along the numerous watercourses that dissect the rich volcanic soils of northeastern NSW. Rapid habitat decline is likely to have fragmented large continuous populations and have significantly diminished local E. grandis numbers.
This study is part of a larger project relying on genetic and ecological research tools to develop adequate management and rehabilitation strategies for rainforest fragments. Newly developed SSR loci were used to investigate if clearing and rainforest fragmentation affected genetic diversity and population connectivity among E. grandis populations in northern NSW. The extensively fragmented nature of the study area (in the Big Scrub and in northern NSW in general) and the temporal scale of the available populations make E. grandis a suitable candidate for genetic studies investigating the consequences of rainforest fragmentation. Data obtained from this and similar research will provide an important platform for in-depth investigations on the correlation between habitat fragmentation and the genetic diversity, gene flow and population fitness of rainforest species.
Materials and methods
Study species and sampling strategy
Elaeocarpaceae is a tropical and warm temperate family including 12 genera. Elaeocarpus is the largest genus with 360 tree species worldwide (24 in Australia) found across all stages of rainforest succession. E. grandis F.Muell. (the Blue Quandong) is a large, fast growing and potentially valuable timber tree commonly found in subtropical and tropical rainforests and distributed from northern NSW to tropical Queensland (Harden, 2000). E. grandis is sometimes synonymized with E. angustifolius, which is geographically widespread, ranging from India to New Caledonia (Coode, 1984). Large trees are strongly buttressed at the base and can grow to over 40 m in height and 2 m in diameter. Leaves, up to 19 cm long, are oblong-elliptical with regularly toothed margins and small domatia, and turn deep red before falling. The flowers are within greenish-white racemes and in late spring produce round blue fruits (up to 3 cm in diameter) that contain hard, deeply sculptured stones protecting the seeds (five locules). In northern NSW, E. grandis is commonly found along alluvial flats and gullies in lowland subtropical rainforest, and is considered to be an early successional species capable of regenerating in available gaps but also persisting in the canopy of mature phase forests.
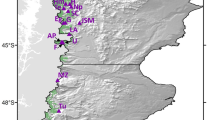
E. grandis was sampled extensively across 21 sites in northern NSW, the species' most southern distribution range (Figure 1, Table 1). These sites were selected according to the size of the E. grandis populations (rather than that of the rainforest remnant) and their distance and isolation from other populations. Two types of populations were sampled: those that had been extensively disturbed and artificially isolated from the continuous forest matrix by clearing (‘fragmented’ populations) and those that were still within the original forest matrix (‘undisturbed’ populations). The fragmented populations were also further divided into the ‘Big Scrub’ group (including populations 5, 6, 11, 12, 13 in Table 1) to represent the most extensively cleared area in the region. One site, Terania Creek, did not fit within either category as it represented a sample from the continuation of the Nightcap NP population within a disturbed matrix, and as a result its data were not included in the analyses concerned with fragmentation. Information on local demographics was gathered at all sites, and either all known individuals were collected (this was the case for all small populations) or sampling reflected the local age structure as accurately as possible. The diameter at breast height (dbh – or the diameter over the buttresses when these were too high) of each individual was measured and all individuals were classified into three groups: old trees (trees >1 m dbh), mature trees (smaller reproductive trees; these were further divided into individuals with dbh over or under 20 cm) and juveniles (saplings and seedlings). For analytical purposes, the plants from each population were also grouped to represent the individuals most likely to be present prior to rainforest clearance and fragmentation (plants >1 m dbh) and those most likely to have germinated after these events (plants <20 cm dbh).
Location of the 21 Elaeocarpus grandis populations sampled in northern NSW for this study. The numbers correspond to those listed in Table 1. Scale bars 50 and 10 km for the main map and inset, respectively.
DNA extraction and SSR analysis
Total genomic DNA was extracted from fresh leaf tissue (or fresh bark if leaves were not accessible) using the DNeasy™ 96 plant kit (Qiagen). The SSR loci used in this study (Table 2) were either selected from those developed by Jones et al (2002) or were previously unpublished (scu31Eg, scu32Eg, scu33Eg, scu34Eg). Polymerase chain reactions (PCRs) contained approximately 5 ng genomic DNA, 0.5 μg BSA, 0.2 μM of each forward and reverse primer (forward primers labelled with either FAM, HEX or NED), 0.2 mM each dNTP and 0.5 U Taq DNA polymerase with reaction buffer (Roche: 10 mM Tris–HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2), in a 12.5 μl reaction. PCR amplifications were performed in a GeneAmp 9700 thermocycler (Perkin-Elmer), using the following conditions: 94°C for 5 min; 35 cycles of 94°C for 30 s, optimum annealing temperature (see Table 2) for 30 s, 72°C for 1 min; and a final extension at 72°C for 5 min. After preliminary verification on 2% agarose, PCR products were separated by capillary electrophoresis on an ABI 3700 Genetic Analyser, and sized by comparison with the Gene Scan 500 (ROX) size standard using the software package Genotyper (version 2.0, Applied Biosystems).
Data analysis
Overall/mean number of alleles and observed/expected heterozygosity were calculated using GDA 1.1 (Lewis and Zaykin, 2002). F and f statistics (Fit and Fis equivalents; Weir and Cockerham, 1984) were calculated using FSTAT 2.9 (Goudet, 1995), with the probability of f being greater than zero determined after 10 000 permutations and 95% bootstrap confidence interval determined after 15 000 bootstraps. Hardy–Weinberg and linkage disequilibrium were assessed through GENEPOP 3.2a (Raymond and Rousset, 1995) using the exact test, and significance levels were determined after 500 batches of 5000 iterations each. Sequential Bonferroni corrections (Rice, 1989) were applied to relevant significance tests. Direct inbreeding values, estimating the loss of heterozygosity over time and based on generational data (Frankham et al, 2002) were also calculated: Fe=1−(Hj/Hm), where Hj is the heterozygosity of the juvenile plants and Hm that of mature individuals.
Information on population size (ie the total number of individuals estimated for the population) and distance from the closest populations and isolation (average distance from other populations) were tested for evidence of correlations with genetic measures of diversity and inbreeding (A, He, Ho, f) using Spearmans rank correlation tests. In order to avoid bias caused by uneven sampling (Leberg, 2002), Rs, a standardised estimate of allelic richness independent of sample size, was calculated for each locus/sample combination using FSTAT 2.9 (Goudet, 1995). The average Rs values across all loci were also used for each population in the correlation tests.
The genetic variance of population structure was determined using RSTCALC 2.2 (Goodman, 1997) to calculate ρ (an unbiased version of the microsatellite-specific measure Rst; Slatkin, 1995) and significance levels were determined after 5000 permutations. The Mantel option in GENEPOP 3.2a was used to test isolation by distance. A matrix of pairwise genetic distances based on δμ2 (Slatkin, 1995) was used as input by NTSYSpc2.11i to produce an UPGMA-based genetic distance tree.
Results
All loci were polymorphic across most populations (only one monomorphic locus/population combination was documented) and produced a total of 39 alleles. Gene diversity, measured as expected heterozygosity, ranged from 0.498 for the only trimeric repeat (Scu32Eg) to 0.724 for Scu33Eg. These loci were also the only two showing significant levels of allelic fixation (Table 2). A possible explanation for low levels of observed heterozygosity can be the presence of null alleles. For instance, locus Scu22Eg (from Jones et al, 2002) was also extensively tested across this study, producing a total of five alleles. However, PCR products from this locus were not amplified across a large number of individuals in the Terania and Nightcap populations, and a preliminary test across 20 seeds from each of five maternal individuals also showed a number of consistently failed amplifications at this locus only (data not presented). As a consequence, Scu22Eg was excluded from the analyses as a cautionary approach, but since such inconsistencies were not observed for Scu32Eg and Scu33Eg these two loci were retained. Pairwise tests of genotypic disequilibrium produced a small number of significant pairings, inconsistent across populations and nonsignificant after sequential Bonferroni corrections.
Table 1 shows the population genetic data summarized across the five loci for the 21 populations investigated (as well as mean values). Allelic diversity across the five loci ranged from 2.4 (Way Way), to 5.0 (Nightcap NP), with a mean value of 3.4. Expected heterozygosity ranged from 0.419 (Alstonville) to 0.683 (Crystal Creek), with a mean value of 0.568. Ho ranged from 0.239 (Maguires Creek) to 0.772 (Crystal Creek), had a mean value of 0.507 and in 15 populations was lower than He. Of the 105 locus/population comparisons, 16 departed from Hardy–Weinberg equilibrium, with the majority (15) showing heterozygosity deficit. Nine populations (six fragmented and three undisturbed) produced f values significantly greater than zero (Table 1). Only the f value for Maguires Creek (0.239) remained significantly positive after sequential Bonferroni corrections; however, it is important to note that increasing evidence suggests that this type of correction can sometimes be far too severe (Devlin et al, 2003).
Overall inbreeding values for E. grandis were f=0.094 (P<0.010) and F=0.173 (P<0.001) and genetic differentiation was ρ=0.076 (P<0.001). Population pairwise comparisons of genetic differentiation ranged between −0.049 and 0.321 (negative values suggest that within-population variance in allele size is bigger than among population variance; Goodman, 1997). The Mantel test did not detect significant correlation (P=0.243; R2=0.009) between genetic differentiation and geographic distance (isolation-by-distance). The UPGMA-based dendrogram in Figure 2 shows the relationship among the 21 populations based on δμ2. Inner Pocket, Rocky Creek and Alstonville form a distinct clade, however neither this nor the other clades produced by this distance measure (or any of the other measures tested; data not presented) correspond to conceivable grouping and/or relationship based on geographic positioning, catchment location or ecological differentiation.
Figure 3 illustrates the relationship between population genetic variables (average allele number, heterozygosity values and inbreeding coefficient) and overall population size, distance from the closest population and, for the fragmented populations, isolation measured as average distance from the other populations. As a general trend, Figure 3 suggests that a greater level of correlation exists between isolation and He, Ho, f than between these values and distance or population size. However, Spearmans rank correlation analysis identified only two significant relationships, the correlations between population size and number of alleles (P=0.004), and between fragment isolation and inbreeding coefficient (P=0.020). The correlation between population size and allelic richness (Rs) was non-significant (R2=0.252, P=0.272), this is not surprising as differences in sampling intensity can bias comparisons of allelic richness among populations (Leberg, 2002). However, it is important to note that the smaller populations were fully sampled; thus, in those instances, lower A represent actual values and not estimates. Overall, Figure 3 displays a trend for larger E. grandis populations to have greater numbers of alleles and for more isolated fragments to have higher inbreeding coefficients.
Relationships between average alleles number, expected heterozygosity, observed heterozygosity, inbreeding and overall population size, distance from closest population and isolation (average distance from the other populations) for the populations used in the fragmentation study. Regression and P-values are indicated.
Adult mature trees represent the greatest proportion of E. grandis plants in northern NSW (61.7%). At undisturbed sites older trees (over 100 cm dbh) represent a considerable proportion (30.8%) of the populations, while juveniles (saplings and seedlings only) are relatively scarce (6.2%, with seedlings missing from half of the sites). Older trees are less common in rainforest fragments (24%), while the proportion of juveniles is marginally higher (7.6%).
Table 3 shows the overall genetic values for populations that are within the natural rainforest habitat, those that are across the fragmented matrix and those that are restricted to the extensively cleared Big Scrub area. Heterozygosity is marginally higher (but not significantly so; Wilcoxon signed rank test), while inbreeding levels are lower within undisturbed populations. The Big Scrub group of populations shows the lowest heterozygosity values. Permutation test (3500 permutations) performed by FSTAT 2.9 between the three groups of populations and based on statistics weighed by sample size (in order to eliminate potential population size bias) failed to identify significant differentiation among these three population groups.
Mean heterozygosity values for all pre-disturbance individuals (ie over 100 cm in dbh) are higher than those for post-disturbance individuals (ie less than 20 cm in dbh), while the opposite is true for inbreeding measures. Such between-generation relationships are generally consistent across all three population groups (Table 3). However, while older plants display similar genetic values across undisturbed, fragmented and Big Scrub populations, there is a trend for loss of heterozygosity (although not significant; Wilcoxon signed rank tests) and increased inbreeding among younger generations in rainforest fragments as compared to undisturbed sites (Table 3). Direct-inbreeding values (Fe) were −0.008, 0.156 and 0.135 for undisturbed, fragmented and Big Scrub groups, respectively.
Discussion
Genetic diversity
This study provides a measure of the genetic diversity present in E. grandis across northern NSW, the species' most southern distribution. Diversity is higher in E. grandis than in two comparative Elaeocarpus species (Rossetto et al, 2004, unpublished), but this was expected as these are restricted in numbers and distribution. No other comparable SSR-based study on Australian rainforest trees has been published, but an extensive study on a non-rainforest outcrossing tree from northern NSW (Melaleuca alternifolia) detected more alleles (98 from five loci) and higher heterozygosity levels (mean He=0.781 and Ho=0.724) from a similar number of individuals sampled across a comparable distribution range (Rossetto et al, 1999).
Similar research on non-Australian rainforest species consistently detected higher levels of genetic diversity. For instance, a greater number of alleles and higher heterozygosity levels were measured among a small number of fragmented sites for Symphonia globulifera (Aldrich et al, 1998) and Carapa guianensis (Dayanandan et al 1999). In studies where heterozygosity levels (Swietenia humilis; White et al, 1999) or number of alleles (Neobalanocarpus heimii; Konuma et al, 2000) were similar to those measured for E. grandis, sampling had been restricted to very narrow ranges. Thus, genetic diversity in E. grandis appears to be higher than in related local endemics but lower than in non-Australian outcrossing rainforest trees. It must be observed, however, that variation in locus-specific levels of polymorphism can introduce bias to cross-species comparisons.
Population structure
Figure 2 and the Mantel test suggest the absence of significant genetic pattern relating to potential ecological or geographic structuring across this species. No meaningful (or reproducible by different distance measures) grouping was produced. Thus, for example, the most geographically distant and disjunct populations, Way Way and Woolgoolga, did not cluster separately from the other sites (Figure 2). Even the apparent loss of heterozygosity of juvenile cohorts within fragmented populations did not affect population structure, with UPGMA-based trees obtained from older, pre-disturbance plants alone displaying the same inconsistencies as Figure 2 (data not presented).
Homogenization of genetic diversity is likely to be a consequence of efficient pollen or seed dispersal mechanisms. Not much is known about Elaeocarpus pollinators, but recent studies suggest that their flowers require buzz pollination (Matthews and Endress, 2002). It could therefore be expected that insects are responsible for the majority of pollen movement in E. grandis. While insect-mediated pollen exchange could be limited (particularly across cleared areas), the fruits produced by E. grandis are consumed by fruit doves and bats (Barker and Vestjens, 1989; Russ, 1999). Since the seeds are protected by a hard endocarp, fruit dispersal is a likely mechanism for genetic exchange and expansion onto suitable habitat. Further studies measuring direct gene flow within and between populations will advance our understanding of the extent and direction of gene flow in E. grandis.
Habitat fragmentation
Over 100 years of logging, clearing and rainforest disturbance have drastically reduced rainforest cover in areas such as the Big Scrub in northern NSW (Lott and Duggin, 1993). Despite the potential re-colonization of a limited number of suitable sites by this early successional species, habitat loss is likely to have led to a rapid decline and fragmentation of previously large and continuous E. grandis, populations. Such drastic changes can be expected to have a significant impact on genetic diversity and population dynamics. For instance, significant losses in diversity have been observed after medium-term disturbance within small Eucalyptus albens remnants (Prober and Brown, 1994), and reduced genetic diversity was measured in Pinus strobus immediately after harvesting (Rajora et al, 2000). In E. grandis, the genetic consequences of habitat fragmentation are not as clear cut as for other species. Table 3 suggests that, on average, undisturbed and fragmented populations do not differ significantly in diversity contents or inbreeding coefficient (as also supported by FSTAT's group tests). There are, however, important trends, albeit not always statistically significant, suggesting that rainforest fragmentation is having an effect. For instance, a greater proportion of fragmented populations show significant inbreeding levels (Table 1), heterozygosity levels decrease with increasing fragmentation (Table 3) and isolation appears to accentuate inbreeding levels (Figure 3).
Dayanandan et al (1999) suggested that the little change observed between fragments and continuous populations of Carapa guianensis in Costa Rica was a consequence of the recent nature of the fragmentation events resulting in the presence of mature trees representing pre-disturbance diversity. Our data show that for early successional species such as E. grandis modest differences are still observed after longer periods since disturbance (between 100 and 150 years). Residual diversity from older plants (which still represent a considerable proportion of all individuals) also appears to influence the overall levels of within-population diversity found in this study. As Table 3 and the Fe values suggest, the proportional loss of heterozygosity among younger cohorts is high in fragmented populations (a likely consequence of isolation), but negative at undisturbed sites.
Another approach for testing the potential influence of overlapping generations on fragmentation data is to assess if increased genetic divergence can be recorded among younger cohorts within isolated populations. For instance, Aldrich et al (1998) reported an increase in genetic divergence among seedlings of Symphonia globulifera following deforestation, which was interpreted as a sign that changes in genetic composition were taking place. In the case of E. grandis, a slight (but nonsignificant) increase in ρ was also detected in younger cohorts within the Big Scrub populations (Table 3), but similar UPGMA-based dendrograms were obtained when analysing older or younger plants separately (data not presented). The latter result, however, is likely to reflect the overall lack of distinguishable population structure in relation to geographic or ecological gradients.
Generally, small isolated fragments can be expected to have a narrow genetic pool, less likely to be replenished. This explains the lower heterozygosity and higher inbreeding levels measured within fragmented populations (particularly across the Big Scrub), as well as the low germination rates of seeds collected from rainforest remnants (Rossetto et al, 2004). It is interesting to note, however, that the rate of genetic decline appears to be more subtle in E. grandis than across other species.
Conclusion
For E. grandis, the genetic consequences of rainforest fragmentation can be measured as a loss of heterozygosity within younger generations. The rate of change appears to be lower than in other species, possibly as the characteristic attributes of early successional species, such as efficient dispersal and fast growth within forest gaps, together with an outcrossed mating system and long life expectation, have inferred a certain degree of resilience to this rainforest tree. Overall, the SSR data also appear to suggest a trend for increased long-term survival of more heterozygous individuals, a matter that requires corroboration through further investigations.
Although this species may be capable of efficiently colonizing new habitat, such a habitat is no longer available because of extensive clearing. The homogeneity of diversity and lack of genetic structure within E. grandis could be explained by post-glacial expansion. However, lowland ‘expansion’ areas (such as the Big Scrub), that during the inter-glacial periods have an important role in facilitating genetic exchange and increases in population size, have now been all but lost to clearing. As a result, when dealing with early successional species, local rainforest restoration projects should aim to replicate inter-glacial expansion events by maximizing local genetic diversity within the material planted.
Future studies on E. grandis will focus on genetic structure across the overall distribution of this species, in order to better understand current and historical radiation patterns. More detailed studies on mating system, dispersal mechanisms, gene flow patterns and correlation between genetic diversity and offspring fitness are also desirable. In the long term, the combination of similar data from a range of taxa will contribute to the development of broadly applicable management and conservation strategies for fragmented rainforests.
References
Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G (1998). Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Mol Ecol 7: 933–944.
Barker RD, Vestjens WJM (1989). The Food of Australian Birds. 1: Non-Passerine. CSIRO Australia: Melbourne.
Bowman DMJS (2000). Australian rainforests – islands of green in a land of fire. Cambridge University Press: Cambridge.
Burbidge NT (1960). The phytogeography of the Australian region. Austr J Bot 8: 75–211.
Connelly S, Specht A (1988). Big Scrub Conservation Strategy. New South Wales National Parks and Wildlife Service: Sydney.
Coode MJE (1984). Elaeocarpus in Australia and New Zealand. Kew Bull 39: 509–586.
Dayanandan S, Dole J, Bawa K, Kesseli R (1999). Population structure delineated with microsatellite markers in fragmented populations of a tropical tree, Carapa guianensis (Meliaceae). Mol Ecol 8: 1585–1592.
Devlin R, Roeder K, Wasserman L (2003). False discovery or missed discovery? Heredity 91: 537–538.
Frankham R, Ballou JD, Briscoe DA (2002). Introduction to Conservation Genetics. Cambridge University Press: Cambridge.
Goodman SJ (1997). RSTCALC: a collection of computer programs for calculating unbiased estimates of genetic differentiation and determining their significance for microsatellite data. Mol Ecol 6: 881–885.
Goudet J (1995). FSTAT, version 1.2: a computer program to calculate F statistics. J Hered 86: 485–486.
Hale ML, Lurz PWW, Shirley MDF, Rushton S, Fuller RM, Wolff K (2001). Impact of landscape management on the genetic structure of red squirrel populations. Science 293: 2246–2247.
Harden GJ (2000). The Flora of New South Wales 2nd edn. New South Wales University Press: Sydney.
Jones RC, McNally J, Rossetto M (2002). Isolation of microsatellite loci from a rainforest tree, Elaeocarpus grandis (Elaeocarpaceae), and amplification across closely related taxa. Mol Ecol Notes 2: 179–181.
Konuma A, Tsumura Y, Lee CT, Lee SL (2000). Estimation of gene flow in the tropical-rainforest tree Neobalanocarpus heimii (Dipterocarpaceae), inferred from paternity analysis. Mol Ecol 9: 1843–1852.
Laurence WL, Bierregaard RO (1997). Tropical Forest Remnants – Ecology, Management, and Conservation of Fragmented Communities. The University of Chicago Press: Chicago.
Leberg PL (2002). Estimating allelic richness: effects of sample size and bottlenecks. Mol Ecol 11: 2445–2449.
Lewis PO, Zaykin D (2002). Genetic Data Analyser: Computer Program for the Analysis of Allelic Data, Version 1.1. Free program distributed by the authors over the internet at http://lewiseeb.unconn.edu/lewishome/.
Lott RH, Duggin JA (1993). Conservation Significance and Long Term Viability of Subtropical Rainforest Remnants of the Big Scrub, North-Eastern New South Wales, Department of Ecosystem Management, University of New England: Armidale.
Matthews ML, Endress PK (2002). Comparative floral structure and systematics in Oxalidales (Oxalidaceae, Connaraceae, Brunelliaceae, Caphalotaceae, Cunoniaceae, Elaeocarpaceae, Tremandraceae). Bot J Linn Soc 140: 321–381.
McDonald WJF, Elsol JA (1984). Moreton Region Map Series: Summary Report and Species Checklist for Caloundra, Brisbane, Beenleigh and Murwillumbah sheets. Queensland Department of Primary Industries: Brisbane.
Prober SM, Brown ADH (1994). Conservation of the grassy white box woodlands: population genetics and fragmentation of Eucalyptus albens. Conserv Biol 8: 1003–1013.
Rajora OP, Rahman MH, Buchert GP, Danick BP (2000). Microsatellite DNA analysis of genetic effects of harvesting in old-growth eastern white pine (Pinus strobus) in Ontario, Canada. Mol Ecol 9: 339–348.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenism. J Hered 86: 248–249.
Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Rossetto M, Gross CL, Jones R, Hunter J (2004). The impact of clonality on an endangered tree (Elaeocarpus williamsianus) in a fragmented rainforest. Biol Conserv 117: 33–39.
Rossetto M, Slade RW, Baverstock PR, Henry RJ, Lee LS (1999). Microsatellite variation and assessment of genetic structure in tea tree (Melaleuca alternifolia – Myrtaceae). Mol Ecol 8: 633–643.
Russ L (1999). Regeneration and flying foxes. In: Horton S (ed) Rainforest Remnants A Decade of Growth. NSW National Parks and Wildlife Service: Hurstville pp 188–206.
Slatkin M (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462.
Smouse PE, Peakall R (1999). Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82: 561–573.
Templeton AR, Shaw K, Routman E, Davis SK (1990). The genetic consequences of habitat fragmentation. Ann Missouri Bot Gard 77: 13–27.
Webb LJ, Tracey JG (1981). Australian rainforests: patterns and change. In: Keast A (ed) Ecological biogeography of Australia. Junk: The Hague pp 605–694.
Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
White G, Boshier DH, Powell W (1999). Genetic variation within a fragmented population of Swietenia humilis Zucc. Mol Ecol 8: 1899–1909.
Whitmore TC (1997). Tropical forest disturbance, disappearance, and species loss. In: Laurence WL, Bierregaard RO (eds) Tropical Forest Remnants – Ecology, Management, and Conservation of Fragmented Communities. The University of Chicago Press: Chicago pp 3–12.
Young A, Boyle T, Brown T (1996). The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11: 413–418.
Young AG, Clarke GM (2000). Genetics, Demography and Viability of Fragmented Populations. Cambridge University Press: Cambridge.
Acknowledgements
This project was funded by an Australian Research Council SPIRT grant (C00107305) and by the NSW National Parks and Wildlife Services. We thank the School of Plant Science at the University of Tasmania for the logistic support given to RJ towards the end of this project, G Hodgson for preparing the map and B Nash and B Horton-Hunter for field assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rossetto, M., Jones, R. & Hunter, J. Genetic effects of rainforest fragmentation in an early successional tree (Elaeocarpus grandis). Heredity 93, 610–618 (2004). https://doi.org/10.1038/sj.hdy.6800585
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800585
Keywords
This article is cited by
-
Small Urban Stands of the Mangrove Avicennia marina are Genetically Diverse but Experience Elevated Inbreeding
Estuaries and Coasts (2015)
-
Genetic structuring of remnant forest patches in an endangered medicinal tree in North-western Ethiopia
BMC Genetics (2014)
-
Contrasting levels of connectivity and localised persistence characterise the latitudinal distribution of a wind-dispersed rainforest canopy tree
Genetica (2014)
-
Fragmentation genetics of rainforest animals: insights from recent studies
Conservation Genetics (2014)
-
Species-site matching in mixed species plantations of native trees in tropical Australia
Agroforestry Systems (2013)