Abstract
In the semienclosed Adriatic Sea, the shared stocks of the cephalopods Loligo vulgaris and Sepia officinalis represent important marine fisheries resources exploited by all coastal countries. The improving of knowledge on the demographic features of these shared stocks is internationally relevant for adopting responsible management and conservation of these marine resources. Analyses of microsatellite variation in geographical samples collected from all parts of the Adriatic Sea were performed using arrays of species-specific di-nucleotide and tri-nucleotide loci. In L. vulgaris the level of genetic variability was consistent with that observed in other loliginid species, whereas the S. officinalis stock showed a microsatellite variation markedly lower than that estimated for the Atlantic and Mediterranean populations collected around the Iberian peninsula. The weak spatial genetic differentiation, the discordant results of the genetic divergence estimators and the lack of any geographical cline in the spatial genetic differences suggest the occurrence of single genetically homogeneous populations within the Adriatic stocks of both species, recommending a coordinated management of the squid and cuttlefish by the Adriatic fishing countries. On the contrary, significant differences detected in temporal replicates of S. officinalis might suggest that allelic frequency can change relating to reproductive behaviour.
Similar content being viewed by others
Introduction
The perception that natural resources are not inexhaustible, the collapse of internationally relevant fisheries stocks and the estimate that 60% of the major marine fisheries are either fully exploited or overexploited (FAO, 1997), promoted a great number of scientific investigations to solve bio-ecological and demographic questions in a huge number of marine populations (Carvalho and Hauser, 1998). The assessment of spatial and temporal patterns of genetic diversity and related demographic features (eg number of migrants, geographical boundaries of populations) may be useful for the conservation and sustainable management of marine resources. Such data can be particularly relevant for the management of stocks whose exploitation is shared by more than one country and which inhabit semienclosed or enclosed basins.
The Adriatic Sea is a semienclosed basin, characterised by an extended continental shelf in the northern and central part and a continental slope in the southern part, where stocks of molluscs, crustaceans, cartilaginous and bony fish are co-exploited by fleets of Italy and Balkan nations. Due to economic and political aspects, the responsible management and conservation of the Adriatic shared fisheries stocks is necessary and needs to be supported by scientific grounds. In 1999, the Food and Agriculture Organization (FAO) implemented the regional Project AdriaMed, whose principal aims are to promote scientific cooperation among Adriatic countries and to improve the management of fishing resources and activities (AdriaMed, 2000). Within the AdriaMed framework, an extensive genetic research project was activated in order to identify population units in the shared stocks of 15 demersal and pelagic fisheries resources that are considered of high priority for the Adriatic fisheries.
The cephalopds European squid (Loligo vulgaris, Loliginidae) and cuttlefish (Sepia officinalis, Sepiidae), share several bio-ecological features, such as a nekto-benthic lifestyle, a relatively short lifespan (12–24 months), reproduction over the whole year at low depths, similar fecundity rates, the absence of a pelagic larval phase and the presence of several cohorts of individuals (Relini et al, 1999, pp 680–686 and 710–714). Weak but relevant differences concern dispersal ability and migratory behaviour. The dispersal ability appears to be greater in L. vulgaris than in S. officinalis because of a more pronounced benthic lifestyle of the latter species, whereas the migratory behaviour is more accentuated in S. officinalis with seasonal coastward migrations of mature individuals for reproduction (Relini et al, 1999, pp 680–686 and 710–714).
In the cephalopods, recent works using microsatellite markers showed genetic homogeneity across the European squid populations occupying the European shelf seas of the NE Atlantic (Shaw et al, 1999), as well in other loliginid species as in the California squid (L. opalescens) populations collected along the North America Pacific coast (Reichow and Smith, 2001). Similarly Adcock et al (1999) found no consistent pattern of geographic or temporal genetic structure in the Southwest Atlantic stock of the squid Illex argentinus. In contrast, a significant population substructuring was found in the cuttlefish samples collected around the Iberian Peninsula. The observed clinal pattern of gene frequencies is consistent with an isolation-by-distance model of low level of gene flow or, alternatively, with a model of secondary contact and introgression between previously isolated populations (Pérez-Losada et al, 2002).
This paper deals with a comparative analysis of spatial and temporal genetic variation patterns on the European squid and cuttlefish samples collected from all parts of the Adriatic Sea. The analysis was carried out using species-specific microsatellite markers (Shaw and Pérez-Losada, 2000; Guarniero et al, 2003) with the aim to assess genetic diversity, population structure, gene flow rates and number of migrants.
Methods
Sampling
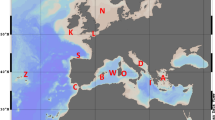
Samples of L. vulgaris and S. officinalis were collected in different locations of the Adriatic Sea (Figure 1) during the MEDITS 2001 scientific trawl survey (Table 1). A sample of S. officinalis (SEOF5) was collected in 2002 from the same location of SEOF4. About 0.5 g of tentacle tissue was dissected using a sterile cutter and stored in 80% ethanol at 4°C.
Geographical locations of cephalopod samples in the Adriatic Sea (see Table 1 for captions).
DNA extraction and microsatellite analysis
Individual genomic DNAs were extracted from 25–50 mg of dried tissue according to a saline procedure (Miller et al, 1988).
The L. vulgaris samples were screened for variation at six dinucleotide microsatellite loci: five were previously developed by Guarniero et al (2003) and one was newly isolated and optimised (locus LVtleB22, Genbank acc. no. AY351669). In S. officinalis, five out of the seven trinucleotide loci developed by Shaw and Pérez-Losada (2000) were used. In both species, the test for linkage disequilibrium between pairs of loci was performed using GENEPOP 3.1 (Raymond and Rousset, 1995) and showed no significant P-values.
The PCR reactions (10 μl total volume) included about 20 ng of genomic DNA, 1.5–2.5 mM MgCl2, 0.5 μM of each primer, 200 μM dNTPs, 10 mM Tris-HCl (pH 9), 50 mM KCl, 0.1% Triton X-100 and 1 U of Taq Polymerase (Promega). PCR reactions were performed on a GeneAMP PCR System 9700 (Applied Biosystems) according to Guarniero et al (2003) for L. vulgaris and to Shaw and Pérez-Losada (2000) for S. officinalis. The locus LVtleB22 was amplified using the primers 5′ → 3′ Forward-AAGTCCTCTATGCCCACTTCC, Reverse-CGATTTCCTCCATGTTGAGC and the following PCR conditions: an initial denaturation at 94°C for 3 min, 35 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final holding at 72°C for 5 min.
The genotyping of individuals was performed by assessing the allele size on an ABI310 automated Genetic Analyser (Applied Biosystems), using forward primers labelled with 6-FAM, HEX or TAMRA (MWG Biotech) and the ROX 400 (Applied Biosystems) as internal standard. Allele sizing was performed using the GeneScan Analysis Software v. 2.02 (Applied Biosystems).
Data analysis
Microsatellite polymorphism within samples was measured as the mean number of alleles per locus, and observed and unbiased expected heterozygosity calculated using the Genetix 4.0 Software package (Belkhir et al, 1996). The deviations from Hardy–Weinberg equilibrium (henceforth HW equilibrium) were tested using the Fisher's exact test with the level of significance determined by a Markov chain method using GENEPOP 3.1 software (Raymond and Rousset, 1995).
The differentiation between samples can be estimated from microsatellite data using statistics based on alternative evolutionary models (ie an infinite allele model, Kimura and Crow, 1964; a stepwise mutation model, Kimura and Ohta, 1978). The most suitable statistical analyses to evaluate the genetic relationships between populations are still a matter of debate (Goodman, 1997; Valsecchi et al, 1997; Gaggiotti et al, 1999; Hardy et al, 2003). For comparative purposes, we chose to use three different methods. First, we tested for simple differentiation in frequencies using the Fisher's exact test implemented in GENEPOP 3.1 Software package (Raymond and Rousset, 1995). Second, we estimated total and pairwise genetic differentiation among samples using FST (Weir and Cockerham, 1984) and using (Goodman's (1997) unbiased RST. We used the F-STAT Software packages (Goudet, 1995) and the RST-CALC program (Goodman, 1997), which adjusts for differences in sample size and allele size variances among loci. Fixation indexes were tested for significant departure from zero using permutation tests available in the respective software packages. Sequential Bonferroni corrections (Rice, 1989) for multiple comparisons were applied where necessary.
To assess whether stepwise mutations affected genetic differentiation in the population, we applied the allele size permutation test (Hardy et al, 2003) implemented in the SPAGeDi software (Hardy and Vekemans, 2002). The principle of this test is to compare the RST value with the distribution of RST values (called pRST) obtained after 1000 random permutations of allele size among allelic states. The observed RST values significantly larger than the pRST indicate that stepwise mutations contributed to the genetic differentiation among subpopulations.
Estimates of the effective number of migrants between sample sites were calculated from the FST values according to Wright's (1978) statistics or using the rare allele method (Slatkin, 1985) implemented in the GENEPOP 3.1 Software (Raymond and Rousset, 1995). The two methods are similar in their sensitivity to detect variation in population structure and both can provide reasonably accurate estimates of Nm under a wide variety of conditions (Slatkin and Barton, 1989).
To test for isolation by distance as an explanation for genetic differences, Mantel tests were performed with GENEPOP (using 1000 permutations to test significance of correlation). The input data for the two matrices were the natural logarithm of the geographical distance (expressed in km) and FST or RST values.
Results
Microsatellite variation in squid and cuttlefish Adriatic samples
Allele sizes and genetic variability at the microsatellite loci in the Adriatic samples of L. vulgaris and S. officinalis are shown in Table 2 and 3, respectively. In L. vulgaris, polymorphism indexes (Na, Ho and He) at the six dinucleotide loci were high and homogeneous over all geographic samples. The Na per locus ranged from 7.5 to 17.7 (average overall loci 12.8). The mean Ho and He ranged from 0.628 to 0.815 (average over all loci 0.682) and from 0.701 to 0.927 (average over all loci 0.837), respectively. The frequencies of the observed heterozygous genotypes were lower than those expected under the HW equilibrium in all the single-locus tests. However, significant deficiencies were observed in four cases out of 24 and only three (12.5%) remained significant after the sequential Bonferroni correction. Significant deviations from HW equilibrium, however, were never associated to any particular locus or sample. At the five trinucleotide loci, the S. officinalis samples showed levels of variability markedly lower than those observed in L. vulgaris. The mean Na, Ho and He ranged from 3.0 to 10.4 (mean 7.4), 0.219 and 0.824 (mean 0.471), 0.351 and 0.891 (mean 0.620), respectively. Despite the low sample size of SEOF2, polymorphism indexes were generally homogeneous (Table 3). Observed heterozygosities were lower than those expected in most single-locus tests and significant deficiencies were found in 15 cases out of 25. After the application of sequential Bonferroni correction, 11 cases (44%) were still significant and the locus Sof1 showed significant departures from HW equilibrium in all samples.
Null alleles are a common problem when using microsatellite loci and they can lead to heterozygote deficiencies (O'Connell and Wright, 1997). Their occurrence may be due either to a mutation in the primer consensus sequence or to technical problems associated with amplification. In our study, re-amplification under less stringent reaction conditions did not change the individual genotypes in both species. In L. vulgaris, the cases with significant departures from HW equilibrium showed the expected frequency of null alleles, calculated as in Brookfield (1996), similar or higher than that the observed one. Thus, null alleles may be invoked to explain significant heterozygous deficiencies. On the contrary, within S. officinalis the expected number of null amplifications was lower than that observed in six out of 11 single locus tests with significant heterozygous deficiency. In this species, the high rate of observed heterozygote deficiency could not be explained only by the occurrence of null alleles.
Population genetic structure of the Adriatic stocks
The exact test for homogeneity of allelic frequencies across all samples showed significant heterogeneity for both L. vulgaris and S. officinalis Adriatic stocks (P<0.01, Table 4). A large proportion (four out of six in L. vulgaris and eight out of 10 in S. officinalis) of significant departures from homogeneity was found in the pairwise comparisons, though none was seen at all loci. In L. vulgaris, the overall estimates of genetic heterogeneity gave contrasting results since nonsignificant and significant genetic heterogeneity values were respectively found using the FST (0.010, not significant) and RST (0.025, P<0.05) estimators. Similarly, significant differences from zero (ie no genetic differences) in the pairwise FST and RST values appeared to be related to the estimator used rather than to a given pair of geographical samples, and none of the comparisons showed significant differences for both indexes. In S. officinalis, the overall FST and RST values calculated including only the spatial samples (SEOF1-SEOF4) were closely similar to those of L. vulgaris (0.011, NS and 0.018, NS, respectively; Table 4). Two out of six pairwise comparisons were significant using FST and one out of six with RST estimates. By contrast, a high number of significant pairwise comparisons involving the SEOF5 temporal sample were found with both estimators (Table 4), including that with the sample SEOF4 collected from the same geographical area.
Since departures from HW equilibrium may affect the estimates of genetic divergence and gene flow among samples, a statistical analysis of microsatellite variation in S. officinalis was successively carried out. In this analysis the locus Sof1 was excluded, since it showed a significant deficiency of heterozygous genotypes in all samples (Table 2). The newly calculated overall values of FST and RST did not change and still remained not significant (0.010, NS and 0.018, NS). The exclusion of the locus Sof1 did not also affect substantially the significance of pairwise comparisons, changing only in the RST values of the comparisons SEOF3–SEOF4 (P<0.01) and SEOF1–SEOF5 (not significant).
In order to assess the efficiency of FST vs RST we applied the allele size permutation test (Hardy et al, 2003) to our data. In both species, the multilocus pRST values did not differ from RST (Table 4) and, at the single-locus level, significant differences between pRST and RST values were detected only in LVrip3 (P<0.01) and Sof4 (P<0.05) loci. Thus, no significant contribution of stepwise mutations to population differentiation for both species was revealed.
The multilocus estimates of the effective number of migrants per generation (Table 4), based on a private allele model (Slatkin, 1985), were 9.7 for L. vulgaris and 7.2 for S. officinalis. These values greatly increased to 24.8 and 16.4 when using overall FST (Wright, 1978). Overall and pairwise Nm estimates indicated high levels of gene flow in samples of both species. In S. officinalis, the removal of the locus Sof1 did not significantly affect the Nm values (Table 4), which was always greater than one for all comparisons. There was no indication for isolation by distance since Mantel tests was not significant in both species.
Discussion
Genetic variability in the Adriatic cephalopod stocks
Polymorphic microsatellite loci are useful genetic markers for indirect monitoring of demographic features of marine fishery resources (Carvalho and Hauser, 1998). In contrast to allozymes, species-specific microsatellites allow more powerful and accurate tests of population genetic parameters in cephalopods (Shaw et al, 1999; Reichow and Smith, 2001; Pérez-Losada et al, 2002). The arrays of microsatellite loci used showed a higher genetic variability in the Adriatic Sea stock of L. vulgaris with respect to S. officinalis. Estimates of genetic variability in the Adriatic stock of the European squid L. vulgaris (overall mean Na 12.8 and He 0.837) are fully consistent with those detected in the NE Atlantic populations of the veneid squid L. forbesi (overall mean Na 11.2 and He 0.78; Shaw et al, 1999) and in the California squid L. opalescens (overall mean Na 12.3 and He 0.825; Reichow and Smith, 2001) using different arrays of di- and trinucleotide loci. On the contrary, the estimates of genetic variability of the Adriatic Sea stock of S. officinalis (overall mean Na 7.4 and He 0.620) were markedly lower than those found in the populations sampled around the Iberian peninsula (overall mean Na 11.5 and He 0.727; Pérez-Losada et al, 2002). In our study, we used the same microsatellite loci used by Pérez-Losada et al (ie loci Sof1, Sof2, Sof4, Sof6 and Sof7), all showing lower Na and He values. There are several possible explanations for such reduced genetic variability, which could be a result of population life history traits (eg bottlenecks) or related to the current population size (eg inbreeding depression). Alternatively, a decrease in both heterozygosity and allele number may indicate severe demographic decline caused by overexploitation of this resources (Hauser et al, 2002). Also, methodological factors such as sample bias or sampling inadequacies may explain the low genetic variability of S. officinalis Adriatic samples. Therefore, further analyses of fine-scaled samples as well temporal replicates will be needed to confirm such genetic feature of the Adriatic cuttlefish stock.
Deficiencies of heterozygous genotypes have been widely detected in cephalopod populations (Shaw and Pérez-Losada, 2000; Pérez-Losada et al, 2002; present study). In the Adriatic L. vulgaris stock, null alleles may be the cause of the observed departures from the HW equilibrium. Design and lab test of new primer pairs will be required to confirm the presence of null alleles due to nucleotide mutations in the primer consensus sequences. Alternatively, a test involving controlled mating experiments to allow pedigree studies might give evidence of null allele occurrence. In the Adriatic cuttlefish, the presence of null alleles cannot completely explain the observed deficiency of heterozygous genotypes, indicating that other bio-ecological factors might affect allele equilibrium in samples. In the Adriatic Sea, the seasonal coastward migrations of mature cuttlefishes for reproduction (Relini et al, 1999, pp 680–686) could cause the admixture of individuals from different cohorts and might contribute to generate the observed departures from HWE in the collected samples (eg Wahlund effect). The mixing of different cohorts over the spawning grounds has been described in cephalopods (Hixon, 1983; Carvalho et al, 1992) and invoked to explain HWE departures (Adcock et al, 1999).
Efficiency of the genetic divergence estimators
Three different estimators of the genetic divergence (ie exact test of sample differentiation, FST, and RST) were used to test the extent of genetic differences within the Adriatic stock of L. vulgaris and S. officinalis. The exact test of differentiation indicated a highly significant overall spatial heterogeneity in sample allelic frequencies (P<0.01) for both L. vulgaris and S. officinalis stocks. However, the single locus test demonstrated that none of the significant comparisons were caused by genetic divergences at all loci and that loci contributed differently to the divergence of samples. The overall estimates of FST and RST gave discordant results in L. vulgaris, showing nonsignificant and significant spatial genetic heterogeneity of samples, respectively. On the contrary in S. officinalis, the overall FST and RST were concordant in revealing a lack of significant genetic heterogeneity within the Adriatic stock. The pairwise FST and RST estimates showed some significant differences for both species, but regarding different pair of samples. In addition, the observed significant differences cannot be related to geography or ecological factors.
On the whole, the level of differentiation found with the exact test was higher than that assessed by the other two estimators. The exact test of sample differentiation gives more weight to rare alleles and can be therefore more sensitive for the detection of weak population differentiation. The most suitable statistical analysis to estimate differentiation among samples at microsatellite loci is still a matter of debate. The RST estimators, based on a stepwise mutation model (Kimura and Ohta, 1978), are usually considered to better predict the type of variation observed at microsatellite loci. After testing a range of statistics, Valsecchi et al (1997) concluded that Goodman's (1997) unbiased RST is probably the most suitable for microsatellite data. On the other hand, Gaggiotti et al (1999) demonstrated that FST estimators based on the infinite allele model are considered more reliable than RST when less than 20 loci are used. O'Connell and Wright (1997) suggested a conservative approach of conventional F-statistics for commercial fish species in the absence of testing and mainly in the species exhibiting a large number of alleles. The allele size permutation test (Hardy et al, 2003) showed that stepwise changes in allelic state do not contribute significantly to the RST estimates, suggesting that we estimated allele frequency changes determined by different genetic effects (eg genetic drift), and that FST should be the more appropriated estimator for assessing genetic differentiation among the Adriatic samples of L. vulgaris and S. officinalis.
Population structure and gene flow
Previous genetic stock structure analysis carried out using microsatellite markers in the veneid squid L. forbesi (Shaw et al, 1999) detected great and highly significant levels of differentiation between samples from Azores and NE Atlantic (FST: 0.255–0.297; RST: 0.222–0.389; P<0.001 for both estimators), suggesting the existence of reproductively isolated populations. The hydro-geographical factors (eg deep water and isolating current regimes) were supposed to act as barriers to the migration and dispersal of squid. Subtle genetic divergences between NE Atlantic samples from shore and shelf waters were suggested by lower but still significant FST and RST values and related to local adaptation in life history characteristics. On the contrary, no significant genetic differences were observed within the continental European shelf stock of veneid squid either in the coastal stocks of the shortfin squid I. argentinus from SW Atlantic (Adcock et al, 1999) or in the California squid L. opalescens (Reichow and Smith, 2001). In these latter cases, both FST and RST estimates were low and effectively zero. The genetic homogeneity shown by coastal and shelf squid stocks was likely related to the lack of hydrographic barriers and to the high dispersal ability of squids.
The lack of a consistent relationship between genetic differences and geographical or ecological factors, the discordant results obtained using different estimators, the low FST and RST values, together with the high Nm estimates, strongly suggest the presence of a genetically homogeneous stock of L. vulgaris in the Adriatic Sea. This issue is consistent with the genetic uniformity observed in the cephalopod stocks inhabiting areas with similar hydro-geographical features (the NE Atlantic stock of L. forbesi, Shaw et al, 1999; the SW Atlantic stock of I. Argentinus, Adcock et al, 1999; the NE Pacific stock of L. opalescens, Reichow and Smith, 2001). If the central-northern Adriatic Sea shows high levels of hydro-geographical homogeneity, the pronounced continental slope of the southern Adriatic could represent a significant geographical barrier for several bottom-associated species living at relatively low depths. The European squid is a nekto-benthic species found in the Italian Seas from the surface to a depth of 425 m, even though it has been proved that the greatest abundance is at depths lower than 100 m (Relini et al, 1999, pp 710–714). Although it is still not clear if adult loliginids can make long-distance migrations through deep water, the genetic similarity observed between the samples from southern Italy and Albania coasts indicates that the south Adriatic continental slope does not reduce gene flow in this area. The Nm estimates among the Adriatic L. vulgaris samples largely exceed those generally considered sufficient to prevent subpopulation structuring (Slatkin and Barton, 1989; Mills and Allendorf, 1996). Therefore, high levels of genetic connectivity are present within the Adriatic squid stock, which can be considered homogeneous across the Adriatic Sea. It is likely that the Adriatic squids belong to a single panmictic population unit whose effective boundaries might be wider than the area investigated. Further analyses using the same set of polymorphic loci and including samples from adjacent Ionian and Aegean Seas are needed to define population boundaries and temporal stability of the genetic homogeneity. After reviewing the genetic features of several stocks in four squid species (Adcock et al, 1999; Shaw et al, 1999; Reichow and Smith, 2001; present paper), it appears that, in these organisms, gene flow should act as the dominant evolutionary force in relatively small geographic areas (ie at the local scale). However, it has also been observed that geographical structuring of populations and restriction of gene flow may exist among samples from wider areas (ie at the regional scale, Shaw et al, 1999).
As in L. vulgaris, low levels of spatial genetic differentiation were detected among the Adriatic samples of S. officinalis. The overall FST and RST estimates were consistently not significant, though some significant pairwise differences were observed, regardless of geographical or ecological factors. The low level of differentiation together with the high gene flow rates among samples argued for the occurrence of a panmictic population in the Adriatic cuttlefish stock. On the contrary, highly significant genetic differentiation was found between samples of S. officinalis collected around the Iberian peninsula, indicating that barriers to gene flow exist among areas (Pérez-Losada et al, 2002). Well-known contemporary hydrographical barriers such as the Almería-Oran oceanographic front in the SW Mediterranean can produce patterns of genetic differences (it presently separate gene pools of other marine species in this area, for example, Zane et al, 2000), even if the genetic microgeographic fragmentation detected in the Iberian cuttlefishes clearly indicated alternative models of genetic differentiation (eg isolation-by-distance, secondary contacts between isolated populations) (Pérez-Losada et al, 2002). The smaller extension of the Adriatic Sea than the area investigated by Pérez-Losada et al (2002) and the lack of pronounced hydro-geographical barriers to the migration of individuals (Relini et al, 1999, pp 9–20) may account for the observed genetic homogeneity in this low-dispersal marine species.
Although the analysis of the temporal variation in allele frequencies included only one replicate collected in a different season, the strong genetic differentiation of SEOF5 detected indicates that Adriatic cuttlefish may undergo temporal genetic changes. In the Adriatic Sea, longitudinal migrations of mature cuttlefishes towards Italian coasts for reproduction have been reported. The spawning period extends throughout the year, with peaks in spring and summer as well as the frequency-size distribution indicated the presence of several cohorts as the result of the extended reproductive period (Relini et al, 1999, pp 680–686). Temporal differences in the allele frequencies within subpopulations were observed in the shortfin squid (I. argentinus, Carvalho et al, 1992), the Atlantic cod (Gadus morhua, Ruzzante et al, 1998) and hake (Merluccius merluccius, Lundy et al, 2000), suggesting that genetically different schools may migrate to spawning grounds at different times within a given area. The presence of genetically different cohorts in the Adriatic stock of S. officinalis may be invoked to explain the genetic divergence observed between temporal replicates in the Southern Adriatic Sea. However, further analysis involving a larger number of samples collected across different years from the same area and in the same season will contribute to resolve the question raised by these preliminary data.
Summarising the data here obtained, the patterns of genetic features detected by the comparative microsatellite variation analysis of the Adriatic stocks of L. vulgaris and S. officinalis suggest the presence of single panmictic populations in both species. This issue recommend a coordinated management of the fisheries stocks by the five Adriatic countries (ie the Republics of Albania, Croatia, Italy, Montenegro and Slovenia). However, since the genetic relationship among cuttlefish samples from different temporal replicates is not yet fully understood, caution regarding fisheries and conservation management of cephalopods should be taken into account.
References
Adcock GJ, Carvalho GR, Rodhouse PG, Shaw PW (1999). Microsatellite analysis of genetic diversity in the squid Illex argentinus during a period of intensive fishing. Mar Ecol Prog Ser 187: 171–178.
AdriaMed (2000) Priority topics related to shared demersal fishery resources of the Adriatic Sea. Report of the First Meeting of the AdriaMed Working Group on Shared Demersal Resources. GCP/RER/010/ITA-02, pp 1–21. Rome: FAO-MiPAF Scientific Cooperation to Support Responsible Fisheries in the Adriatic Sea, http://www.faoadriamed.org/html/av_doc1.asp?id=7.
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996). GENETIX 4.04, logiciel sous Windows TM pour la génétique des populations Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France).
Brookfield JF (1996). A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5: 453–455.
Carvalho GR, Hauser L (1998). Advances in the molecular analysis of fish population structure. Ital J Zool 65(Suppl 1): 21–33.
Carvalho GR, Thompson A, Stoner AL (1992). Genetic diversity and population differentiation of the shortfin squid Illex argentinus in the south-west Atlantic. J Exp Mar Biol Ecol 158: 105–121.
FAO (1997) In: Review of the State of the World Fishery Resources: Marine Fisheries Food and Agriculture Organization, Rome, FAO Fisheries Circular 920 FIRM/C920.
Gaggiotti OE, Lange O, Rassmann K, Gliddon C (1999). A comparison of two indirect methods for estimating average levels of gene flow using microsatellite data. Mol Ecol 8: 1513–1520.
Goodman SJ (1997). RST CALC: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and a determining their significance. Mol Ecol 6: 881–885.
Goudet J (1995). Fstat version 1.2. A computer program to calculate F-statistics. J Hered 86: 485–486.
Guarniero I, Garoia F, di Placido R, Ramsak A, Mannini P, Tinti F (2003). Species-specific microsatellite loci for the European squid (Loligo vulgaris). Mol Ecol Notes 3: 312–313.
Hardy OJ, Charbonnel N, Freville H, Heuertz M (2003). Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163: 1467–1482.
Hardy OJ, Vekemans X (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Hauser L, Adcock GJ, Smith PJ, Bernal Ramirez J, Carvalho GR (2002). Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper. Proc Natl Acad Sci USA 99: 11742–11747.
Hixon RF (1983). Loligo opalescens. In: Boyle PR (ed) Cephalopod Life Cycles, Vol 1: Species Accounts. Academic Press: London, pp 95–114.
Kimura M, Crow JF (1964). The number of alleles that can be maintained in a finite population. Genetics 8: 725–738.
Kimura M, Ohta T (1978). Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc Natl Acad Sci USA 75: 2868–2872.
Lundy CJ, Rico C, Hewitt GM (2000). Temporal and spatial genetic variation in spawning grounds of European hake (Merluccius merluccius) in the Bay of Biscay. Mol Ecol 9: 2067–2079.
Miller SA, Dykes DD, Polesky HF (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16: 1215.
Mills LS, Allendorf FW (1996). The one-migrant-per-generation rule in conservation and management. Conserv Biol 10: 1509–1518.
O'Connell M, Wright JM (1997). Microsatellite DNA in fishes. Rev Fish Biol Fish 7: 331–363.
Pérez-Losada M, Guerra A, Carvalho GR, Sanjuan A, Shaw PW (2002). Extensive population subdivision of the cuttlefish Sepia officinalis (Mollusca: Cephalopoda) around the Iberian Peninsula indicated by microsatellite DNA variation. Heredity 89: 417–424.
Raymond M, Rousset F (1995). Genepop (Version 1.2.): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Reichow D, Smith MJ (2001). Microsatellites reveal high levels of gene flow among populations of the California squid Loligo opalescens. Mol Ecol 10: 1101–1109.
Relini G, Bertrand J, Zamboni A (eds) (1999). Synthesis of the knowledge on bottom fishery resources in central Mediterranean (Italy and Corsica). Biol Mar Mediterranea 6(Suppl 1): 1–868.
Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Ruzzante DE, Taggart CT, Cook D (1998). A nuclear DNA basis for shelf- and bank-scale population structure in northwest Atlantic cod (Gadus morhua): Labrador to Georges Bank. Mol Ecol 7: 1663–1680.
Shaw PW, Pérez-Losada M (2000). Polymorphic microsatellites in the common cuttlefish Sepia officinalis (Cephalopoda). Mol Ecol 9: 237–238.
Shaw PW, Pierce GJ, Boyle PR (1999). Subtle population structuring within a highly vagile marine invertebrate, the veined squid Loligo forbesi, demonstrated with microsatellite DNA markers. Mol Ecol 8: 407–417.
Slatkin M (1985). Rare alleles as indicators of gene flow. Evolution 39: 53–65.
Slatkin M, Barton NH (1989). A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349–1368.
Valsecchi E, Palsboll P, Hale P, Glockner-Ferrari D, Ferrari M, Clapham P et al (1997). Microsatellite genetic distances between oceanic populations of the humpback whale (Megaptera novaeangliae). Mol Biol Evol 14: 355–362.
Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Wright S (1978). Evolution and Genetics of Populations: Variability Within and Among Natural Populations. Vol 4, University of Chicago press: Chicago.
Zane L, Ostellari L, Maccatrozzo L, Bargelloni L, Cuzin-Roudy J, Buchholz F, Patarnello T (2000). Genetic differentiation in a pelagic crustacean (Meganyctiphanes norvegica: Euphausiacea) from the North East Atlantic and the Mediterranean Sea. Mar Biol 136: 191–199.
Acknowledgements
We thank the managing editor and the anonymous referee for helpful comments and criticisms that improved the final version of the manuscript. This study and the fellowship to AR were supported by the AdriaMed Research Project ‘Identification of population units in the Adriatic shared stocks by genetic structure analysis’ granted by FAO-AdriaMed. FG, IG, and ML were supported by research fellowships granted by the Italian Ministry of Agricultural and Forestry Policies. The lab work of this study was carried out at the National Institute of Biology of Ljubljana and at the GenMAP lab of the University of Bologna in Ravenna.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Garoia, F., Guarniero, I., Ramšak, A. et al. Microsatellite DNA variation reveals high gene flow and panmictic populations in the Adriatic shared stocks of the European squid and cuttlefish (Cephalopoda). Heredity 93, 166–174 (2004). https://doi.org/10.1038/sj.hdy.6800489
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800489
Keywords
This article is cited by
-
Genome-wide SNPs reveal complex fine scale population structure in the California market squid fishery (Doryteuthis opalescens)
Conservation Genetics (2021)
-
Omics approaches for conservation biology research on the bivalve Chamelea gallina
Scientific Reports (2020)
-
Genetic population structure and phylogeny of the common octopus Octopus vulgaris Cuvier, 1797 in the western Mediterranean Sea through nuclear and mitochondrial markers
Hydrobiologia (2018)
-
Population genomics of an endemic Mediterranean fish: differentiation by fine scale dispersal and adaptation
Scientific Reports (2017)
-
Microsatellite DNA variation of oval squid Sepioteuthis sp. 2 reveals a single fishery stock on the coastline of mainland Japan
Fisheries Science (2015)