Abstract
Genetic information has played an important role in the development of management units by focusing attention on the evolutionary properties and genetics of populations. Wildlife authorities cannot hope to manage species effectively without knowledge of geographical boundaries and demic structure. The present investigation provides an analysis of mitochondiral DNA and microsatellite data, which is used to infer both historical and contemporary patterns of population structuring and dispersal in the eastern grey kangaroo (Macropus giganteus) in Australia. The average level of genetic variation across sample locations was one of the highest observed for marsupials (h=0.95, HE=0.82). Contrary to ecological studies, both genic and genotypic analyses reveal weak genetic structure of populations, where high levels of dispersal may be inferred up to 230 km. The movement of individuals was predominantly male-biased (average Nem=22.61, average Nfm=2.73). However, neither sex showed significant isolation by distance. On a continental scale, there was strong genetic differentiation and phylogeographic distinction between southern (TAS, VIC and NSW) and northern (QLD) populations, indicating a current and/or historical restriction of gene flow. In addition, it is evident that northern populations are historically more recent, and were derived from a small number of southern founders. Phylogenetic comparisons between M. g. giganteus and M. g. tasmaniensis indicated that the current taxonomic status of these subspecies should be revised as there was a lack of genetic differentiation between the populations sampled.
Similar content being viewed by others
Introduction
Many native species pose a difficult challenge for wildlife managers, and lead to a dichotomy in population management strategies. Many Australian marsupial species have declined because of habitat loss and fragmentation, competition from introduced herbivores and predation by introduced carnivores, while many others have become locally overabundant. Regardless of whether a species is considered as a natural resource to be conserved or a pest to be controlled, a species cannot be managed effectively without knowledge of its biological boundaries and demic structure. Traditional ecological approaches for establishing population management strategies use intensive fieldwork and monitoring of marked individuals to provide detailed information on current demography, rate of recruitment and movement. However, there are significant limitations associated with these approaches (Moritz and Lavery, 1996): (1) ecological studies are usually a snapshot in time, and usually do not encompass stochastic environmental effects that have significant long-lasting effects on community organization; (2) ecological studies are often very limited in space, because they are focused on a small part of the range of the species or community and (3) ecological studies based on marked individuals may be restricted to specific age, sex or size classes to which accessibility is the easiest.
The recent application of high-resolution genetic markers has proven to be a highly effective tool in the investigation of evolutionary processes and population dynamics within species (Sunnucks, 2000). While traditional ecological studies have yielded invaluable insights into behavioral patterns in animal populations, molecular markers have advanced our ability to identify new information about the biology of the species including; genetic structure, dispersal, social structure and identification of individuals within unique populations.
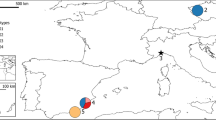
The eastern grey kangaroo (Macropus giganteus) was one of the first kangaroo species to be described by members of Captain James Cook's expedition in 1770 (Frith and Calaby, 1969). M. giganteus is also one of the largest and most commonly observed macropod species, having a wide and almost continuous distribution down the east coast of Australia, where annual rainfall is more than 250 mm (Figure 1; Poole, 1982). Two subspecies are recognized, based on differences in skull morphology and pelage; M. g. giganteus from mainland Australia (QLD, NSW, SA and VIC), and M. g. tasmaniensis from northeast Tasmania (TAS) (Figure 1; Kirsch and Poole, 1972). Although these subspecies have been distinguished by morphological differences, no genetic data exist to test these classifications.
Map of Australia showing geographical distribution (from Poole, 1995) and sampling locations of M. giganteus. Populations within ‘box’ signify samples used to determine fine-scale population structure and dispersal. Sample locations identified by a ‘filled circle’ constitute populations used to determine broad-scale population genetic differentiation (refer to Table 1 for sample sizes). Sample localities represented by letters constitute additional individuals used in mtDNA variability and phylogeography analysis. Sample sizes for these are; a=1, b=1, c=2, d=2, e=1, f=1, g=2, h=1, i=1, j=1, k=2, l=1, m=2, n=1, o=6, p=3 and q=10.
Group home ranges have been identified from a handful of ecological studies for M. giganteus (Kaufmann, 1975; Jarman and Taylor, 1983; Johnson, 1989; Jaremovic and Croft, 1991), and comprehensive information of dispersal is sketchy and unbalanced. However, from these limited studies, females have demonstrated strong site fidelity where they tend to live out their lives close to female relatives, while males range widely with observed range length increasing with the size of the animal, probably reflecting their feeding needs or their greater reproductive activity. In general, these data suggest that both sexes are relatively sedentary, with only a few individuals dispersing distances up to 17 km.
M. giganteus groups have been shown to vary in size and sex/age composition, throughout different parts of Australia (Jarman and Taylor, 1983; Johnson, 1989; Jaremovic and Croft, 1991), which is partly related to habitat structure and resource availability. Populations have now become overabundant in many parts of Australia including rural areas and nature reserves (Hill et al, 1988; Coulson, 1998; Coulson et al, 1999).
Management of wildlife populations can be divided into three areas: pest control, conservation and sustained yield harvesting (Caughley and Sinclair, 1994). Whichever is the case, the management of M. giganteus requires population monitoring and a comprehensive understanding of the biology of the animal. This project aims to provide analysis of genetic data, which will be used to infer both historical and contemporary patterns of population structuring and dispersal.
Methods
Sample collection and DNA extraction
Detailed information on contemporary population structure and dispersal patterns was obtained using six southern New South Wales (NSW) populations sampled at increasing distances. In addition, individuals from two localities within southern Queensland (QLD) were also sampled to evaluate broad-scale population genetic differentiation (Figure 1). At each location, animals were sampled within a discrete area of approximately 3 km2, which typically contained several social groups. To investigate species diversity and phylogeography, additional M. giganteus ear tissue samples were opportunistically obtained from 24 localities throughout the range of M. g. giganteus and from one population of M. g. tasmaniensis (Figure 1). DNA extractions were carried out according to the ‘salting-out’ procedure described in Sunnucks and Hales (1996).
Microsatellite amplification and screening
Microsatellite genetic diversity was assessed using 10 heterologous macropodid loci derived from the tammar wallaby (M. eugenii), Me14, Me17, Me28, T19-1 and T46-5 (Taylor and Cooper, 1998; Zenger and Cooper, 2001a); allied rock-wallaby (Petrogale assimilis), Pa55, Pa297 and Pa597 (Spencer et al, 1995; Spencer, 1996) and yellow-footed rock-wallaby (P. xanthopus), Y148 and Y151 (Pope et al, 1996). Microsatellite loci were amplified via PCR in 10 μl reaction volumes containing; 100–200 ng of genomic DNA, 2.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.1% Triton X-100, 0.1% Tween 20 and NP40, 200 μM each of dCTP, dGTP and dTTP, 20 μM dATP, 0.05 μl of [α33-P]dATP at 1000 Ci/mmol, 1.0 μM of each primer and 0.5 U Taq polymerase (Qiagen). PCR amplifications were carried out using an MJ Research PTC100 thermocycler, with an initial 94°C denaturation for 3 min, followed by ‘touchdown’ cycles of 94°C denaturation for 30 s, annealing temperatures (60, 58, 56, 54, 52 and 50°C) for 45 s and an extension step of 72°C for 1 min. On completion of the last touchdown cycle another 30 cycles were performed at 50°C annealing temperature with a final extension of 72°C for 5 min. The amplified PCR products were resolved on 6% denaturing polyacrylamide gels with a standardized DNA size reference marker and visualized by autoradiography according to Taylor et al (1994).
Mitochondrial DNA amplification and screening
Mitochondrial DNA (mtDNA) control region variation was revealed using the single-stranded conformation polymorphism (SSCP) technique (Sunnucks et al, 2000). The mtDNA control region was amplified by PCR using marsupial-specific primers (mt15999L and mt16498R; Fumagalli et al, 1997). PCR reactions were carried out as described above, except that a single annealing temperature of 60°C was used. The amplified PCR products were resolved on a nondenaturing polyacrylamide gel according to Sunnucks et al (2000). To investigate sequence differences among the unique mtDNA haplotypes, new PCR products (devoid of [α33-P]dATP) from three independent individuals for each identified haplotype were purified, sequenced using BigDye termination (Perkin-Elmer Applied Biosystems) and resolved on an ABI 377 sequencer. In addition, three western grey kangaroo (M. fuliginosus) individuals were sequenced to provide a comparative outgroup in the phylogenetic analysis. All resultant sequences have been deposited in GenBank under accession numbers AF443122 to AF443175.
Diversity indices
Allelic diversity (A) and heterozygosity (observed HO and Hardy–Weinberg expected HE) of the microsatellite loci were estimated using the BIOSYS program (Swofford and Selander, 1981). Exact tests for Hardy–Weinberg equilibrium and genotypic linkage disequilibrium were calculated across all microsatellite loci using the Markov chain method with 1000 iterations calculated in GENEPOP version 3.1 (Raymond and Rousset, 1995). The level of inbreeding and/or the presence of null alleles were assessed by measuring FIS and its significance (10 000 permutations) for all loci within all populations using FSTAT version 2.9.1 (Goudet, 2000). When performing multiple simultaneous comparisons, the sequential Bonferroni procedure (Rice, 1989) with α=0.05 was used to adjust the statistical significance level.
Estimates of mtDNA haplotypic diversity (h) and nucleotide diversity (π) within populations and nucleotide divergence (dA) among populations were calculated using the software package REAP (McElroy et al, 1991). Divergence at the sequence level was calculated using the Kimura 2-parameter (K2P) genetic distance (Kimura, 1980) with a gamma distribution value of α=0.5 (Wakeley, 1993) within MEGA version 2.1 (Kumar et al, 2001). Evidence for population expansion was evaluated using the mismatch distribution analysis (Schneider and Excoffier, 1999) calculated in ARLEQUIN version 2.0 (Schneider et al, 1997).
Population structure
Levels of genetic structure derived from microsatellite data were evaluated using both genic and genotypic methods. Firstly, pairwise values of θST (an unbiased estimator of FST) and their significance were calculated using FSTAT version 2.9.1 (Goudet, 2000), incorporating 10 000 permutations. Secondly, mutilocus genotype data were used to infer population structure via a model-based Bayesian clustering method implemented within the STRUCTURE software package of Pritchard et al (2000). The results presented are based on a modeling approach to infer the number of populations (K), deduced by posterior probability using 106 iterations following a burn-in period of 30 000 simulations. Finally, overall and sex-specific pairwise relatedness values were compared within and among populations to investigate contemporary fine-scale structuring and sex-biased dispersal. All possible pairwise relatedness values were calculated using the relatedness coefficient (R) within the program KINSHIP version 1.2 (Goodnight et al, 1998). The significance of within- versus among population relatedness and male versus female relatedness was calculated using a resampling statistic (10 000 permutations), performed within the program RESAMPLING STATS version 5.0.2 (Simon, 1999).
Patterns of mtDNA geographical structuring were examined using two approaches. Firstly, pairwise ΦST values (K2P with gamma at 0.5) were calculated with associated significant levels (10 000 permutations) across all populations using the ARLEQUIN version 2.0 package (Schneider et al, 1997). Secondly, a nested hierarchical analysis of genetic differentiation was calculated using the AMOVA application (Excoffier et al, 1992) performed within the ARLEQUIN version 2.0 package (Schneider et al, 1997). This program estimates the proportion of total genetic variation within and among the different hierarchical levels based on distribution of haplotypes and pairwise distances. This analysis was calculated using three models: (i) NSW populations as one group; (ii) QLD populations as one group and (iii) NSW and QLD populations combined as two groups.
Gene flow and geographic distance
Pairwise distance values were used to estimate the number of individuals that migrate between each pair of sampling locations per generation. The relations Nem=[(1/θST)−1]/4 was used to estimate the net migration rates using microsatellite nuclear markers (Wright, 1951), while Nfm=[(1/ΦST)−1]/2 was used to investigate the relative amount of female migration using mtDNA data (Slatkin, 1993; Baker et al, 1994).
Analyses of correlation between gene flow and geographic distances were carried out using the Mantel (1967) permutation test executed by the ISOLDE routine (10 000 permutations) in GENEPOP 3.1 (Raymond and Rousset, 1995). All pairwise genetic distance indices including microsatellite θST, mtDNA ΦST and mean relatedness values (R) (male and female) were compared with geographical distance (km) between localities.
Phylogenetic analysis
Evolutionary relationships among the mtDNA haplotypes were inferred using both Neighbor-Joining (NJ) distance and Maximum-Parsimony (MP) methods for phylogenetic reconstruction calculated in PAUP 4.0b8 (Swofford, 2000). The NJ distance method used the genetic distance as previously described (see Diversity indices), while a heuristic search with gaps identified as a fifth state was utilized in the MP method. Statistical support for the branching topology was estimated by bootstrap replications of 1000 for both methods. Both methods used M. fuliginosus sister taxa sequences as the outgroup.
Results
Microsatellite genetic variability
A total of 171 individuals (76 males and 95 females) were genotyped from eight localities (Table 1) using the 10 microsatellite loci. All of the loci were highly polymorphic and exhibited eight to 23 alleles. Two of the loci (Me14 and Y151) indicated the presence of null alleles during genotyping (ie, faint and missing bands). This was verified by large positive FIS values (0.129 and 0.130 respectively) that deviated significantly (P<0.001) from Hardy–Weinberg equilibrium. Both of these loci were subsequently removed from the remainder of the analysis. There was no evidence of linkage disequilibrium between any of the loci.
Allelic diversity (A) and heterozygosity (HO and HE) across the eight remaining microsatellite loci averaged 5.6–10.4 alleles per locus, with observed heterozygosity ranging from 0.766 to 0.852 (Table 1). Under Hardy–Weinberg equilibrium only Marulan demonstrated a significant departure (P<0.05) with a positive FIS value (0.078), indicating a mild Wahlund effect (Table 1).
mt DNA variability
Estimates of mtDNA diversity within and between populations were derived from eight localities incorporating between 10 and 48 individuals (Table 1). Haplotypic diversity (h) and nucleotide diversity (π) within populations averaged 76.78 and 1.76%, respectively (Table 1). Nucleotide divergence (dA) among populations ranged between 0.06 and 3.85%. Hierarchical comparisons revealed that the greatest nucleotide divergence occurs between geographical regions (NSW and QLD; mean=2.90%, SD=0.53%), while comparisons within regions demonstrated the least divergence (mean=0.46%, SD=0.35%). On average, there was five times the amount of nucleotide diversity within populations as among populations in the same geographic region.
Divergence at the sequence level was derived from 51 haplotypes obtained from 202 individuals sampled across the range of M. giganteus (Figure 1). Of 648 bp of sequence, 81 sites (12.5%) were variable, 68 of which were parsimony informative (Figure 2). Haplotypic diversity (h±SD) across the species range was extremely high (94.83±0.66%). Sequence divergence between all haplotypes varied from 0.16% to 6.88% with a mean (±SE) of 3.33±0.50%. When comparing subspecies, there was more sequence divergence within M. g. giganteus (mean 3.36±0.50%), than between the taxa (mean 2.99±0.53%). M. g. giganteus haplotypes within southern Australia (NSW and VIC) demonstrated the greatest mean sequence divergence (3.23±0.48%), while haplotypes within northern Australia (QLD) shared the least (0.86±0.17%). The mean sequence divergence between the two regions was high (4.49±0.74%). Mean sequence divergence between M. giganteus and M. fuliginosus was 14.96±1.97%.
Variable sites of a 648-bp segment of mtDNA control region from 202 eastern grey kangaroos. Variable nucleotide positions are relative to the beginning of the sequence. Dots represent identical bases to the first haplotype. Haplotypes are labeled relative to their geographical region, that is, ‘northern’ (eg, N1), ‘southern’ (eg, S1) or ‘Tasmanian’ (eg, T1).
Population structure
Both pairwise θST and ΦST values revealed significant differences (P<0.05) between populations both before and after Bonferroni correction, with θST results varying between –0.0037 and 0.0967 and ΦST values ranging from 0.0395 to 0.8511 (Table 2). Not surprisingly, comparisons between QLD and NSW populations produced the greatest differences, while pairwise comparisons involving Marulan and N. Bathurst on average demonstrated the least significant differences (Table 2).
The model-based clustering method, for inferring the number of populations (K) based on posterior probability (Pritchard et al, 2000), indicated that samples from NSW and QLD could only be assigned to two separate clusters (P=0.999). When analyzing each of these groups separately, both produced clustering patterns consistent with a single population (P=0.999). For both analyses, all other values of K were deemed highly improbable (P<0.0001). Consequently, no structure could be inferred among the sampled populations within NSW or QLD.
Analysis of mtDNA differentiation using the nested analysis of variance approach (Excoffier et al, 1992) revealed that 49% of total molecular variance (ΦCT=0.486; P<0.05) was partitioned between NSW and QLD when analyzed together as separate groups. In addition, the analysis distributes 40% of the variation (ΦST=0.594; P<0.01) within populations, while the remaining 11% was among populations within groups (ΦSC=0.211; P<0.01). When analyzing NSW and QLD groups separately, the majority (>79%) of the variation was distributed within populations (ΦST=0.214 and 0.148, respectively; P<0.01).
Based on the differentiation between NSW and QLD populations and the small number of sites sampled in QLD, the remaining genotypic analyses were confined to the six NSW populations. Female pairwise relatedness analysis demonstrated a marginal but significant (P<0.001) greater mean within-population relatedness when compared to mean among-population relatedness, while males demonstrated none (P=0.212; Figure 3). Although it is evident that females have a higher within-population relatedness values than males (0.042 and 0.026 respectively), there was no significant difference between them (P=0.402). In general, mild structuring was only evident for females, with all mean relatedness values only slightly deviating from zero (Figure 3). These results are suggestive of females having higher site fidelity.
Relation between pairwise R and corresponding distance values for male (a) and female (b) eastern grey kangaroos. Within-population values indicated by open diamonds, while among-population values are denoted by filled diamonds. Mean pairwise relatedness (±SE) within populations and among populations are 0.026±0.0005, −0.019±0.0004 for males and 0.042±0.0005, −0.007±0.0004 for females. Solid line represents isolation by distance correlation. Broken line indicates average within- versus among-population differences.
Gene flow and geographical distance
The average number of migrants (Nem) among NSW populations was high at 22.61 individuals per generation, with a range of 8.17–59.30 (excluding infinity; Table 2). When examining the average number of female immigrants per generation within NSW, Nfm was 2.73 with a range of 0.60–12.16 (Table 2). Although females demonstrate smaller migration rates compared to the sexes combined, the values are still comparatively high. These results are consistent with previous data, which suggest little genetic differentiation and sex-biased dispersal among M. giganteus populations. As expected, comparisons between NSW and QLD populations produced the smallest values (Table 2).
Analysis of isolation by distance across NSW using the Mantel test, revealed no significant relation between any of the three pairwise genetic distance indices (θST, ΦST and R) and geographical distance (P=0.25, 0.88 and 0.74, respectively). When comparing pairwise relatedness values separately between the sexes, there was again no detectable association (males: P=0.75, females: P=0.74). In fact, populations separated by large distances (∼230 km) demonstrated equivalent numbers of close relatives when compared to populations separated by small distances (∼20 km; Figure 3).
Phylogeographical analysis
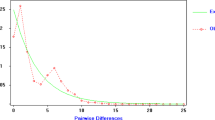
The reconstruction of relations between M. giganteus haplotypes using parsimony and NJ methods produced trees with corresponding topologies in relation to both the positioning and support of the groups (Figure 4). Phylogenetic relations between the subspecies revealed that M. g. tasmaniensis haplotypes were closely related to some M. g. giganteus haplotypes and clustered within one of the four major southern mainland lineages. Within M. g. giganteus, northern Australian (QLD) haplotypes were monophyletic (98–99% support), while there were four main lineages with significant bootstrap support (>80%) for southern Australian haplotypes (NSW and VIC). Two of these lineages, however, cluster with and are basal to the northern lineage (83–94% support), making the mtDNA of southern M. giganteus paraphyletic. There was no obvious structuring within both the northern and southern populations, as haplotypes from different lineages were spread throughout the different populations (Figure 4; Table 2). Furthermore, northern haplotypes exhibit reduced divergence compared to southern lineages, suggesting a recent derivation from southern stock. This claim is also supported by the mismatch analysis which indicates a historical population expansion (SSD P-value=0.49).
NJ tree displaying relation between eastern grey kangaroo mtDNA haplotypes and geographic region. M. g. tasmaniensis haplotypes identified by the arrow, while all other haplotypes are M. g. giganteus except for the outgroup which are M. fuliginosus (western grey eg, WG1) individuals. Values at nodes indicate the statistical support as obtained from 1000 bootstrap replicates. Numbers in plain text are from NJ analysis, while values in bold are derived from the MP method.
Discussion
Genetic diversity
The eastern grey kangaroo has extensive genetic diversity in both microsatellite and mtDNA variability. The mean number (±SD) of observed alleles across the 10 microsatellite loci was 15.8±5.1. Unfortunately, two of the heterologous loci (Me14 and Y151) displayed significant frequencies of null alleles, and were subsequently removed. Recently, species-specific microsatellite loci have been developed for M. giganteus to alleviate this problem (Zenger and Cooper, 2001b). Utilizing the remaining eight microsatellite loci, the average expected heterozygosity (HE±SD) across all populations was high at 0.82±0.02 (Table 1). This level is one of the highest recorded for marsupial taxa (HE range 0.05–0.86, n=24; see Bowyer et al, 2002). Haplotypic diversity (h±SD) across all M. giganteus individuals was also high (94.83±0.66%), and again greater than most other marsupial taxa (h range 12–99%, n=14; see Bowyer et al, 2002). Based on these data, it appears that M. giganteus has had a relatively large long-term effective population size (Frankham, 1996).
The sequence divergence between M. giganteus mtDNA haplotypes varied between 0.16 and 6.88% and is within the range observed from other macropods including the red kangaroo, M. rufus (0.1–6.2%; Clegg et al, 1998) and P. xanthopus (0.3–7.3%; Pope et al, 1996). Surprisingly, there was greater average sequence divergence within M. g. giganteus than between the subspecies, indicating that M. g. giganteus and M. g. tasmaniensis are not well differentiated genetically. In fact, the minimum sequence divergence between the taxa was only 0.47% (three substitutions; Figure 2). This value is substantially less than the minimum differences observed between P. xanthopus subspecies (5.20%; Pope et al, 1996), and between mainland and Tasmanian tiger quolls, Dasyurus maculatus (2.71%; Firestone et al, 1999).
When comparing geographical regions, haplotypes from southern Australia (NSW and VIC) demonstrated greater mean sequence divergence (3.23%) than those from northern Australia (QLD; 0.86%), suggesting a recent common origin for the QLD lineage of M. g. giganteus (see Phylogeography). Although M. g. giganteus is characterized by high mtDNA genetic diversity, the variation within each geographic region (NSW and QLD) is mostly distributed within (π=1.76%) rather than among populations (dA=0.46%). These values are indicative of weak genetic structure (see below), which has also been demonstrated in M. rufus (π=2.90%, dA=0.70%; Clegg et al, 1998).
Population structure and dispersal
Although there are limited ecological data for dispersal patterns in M. giganteus (eg, Kaufmann, 1975; Jarman and Taylor, 1983; Jaremovic and Croft, 1991), these studies suggest strong site fidelity, whereby individuals only disperse small distances (<20 km). Unfortunately, these conclusions may be limited by their methodology (see Introduction), resulting in an incomplete representation of dispersal for this species.
Contrary to these ecological studies, our results reveal weak genetic structure of populations both on a local (<50 km) and regional scale (50–230 km), from which high levels of dispersal may be inferred. Although several pairwise θST and ΦST values were significant (Table 2), both distance indices only revealed relatively small amounts of differentiation between the populations at these spatial scales (mean=0.015, 0.183, respectively). Similarly, only 21% of total mtDNA variation was distributed among NSW populations when analyzed using AMOVA (Excoffier et al, 1992). Further analysis using the multilocus clustering method of Pritchard et al (2000) revealed comparable results, whereby no genetic subdivision could be resolved within this group and the six NSW populations were considered to function as a single population.
Population processes, such as individual movements and relatedness of interacting individuals, were investigated using genotypic approaches to identify fine-scale dispersal of individuals within NSW. Pairwise relatedness analysis indicated weak genetic structuring, whereby individuals are dispersing and breeding across all populations. In particular, females demonstrated only slightly higher levels of site philopatry than males. This sex-biased dispersal is also evident when comparing the average number of migrants (Nem=22.61) to the average number of female migrants (Nfm=2.73) among NSW populations. These results are consistent with numerous M. giganteus ecological studies that suggest greater male dispersal associated with breeding and social structure constraints (eg, Johnson, 1989; Jaremovic and Croft, 1991). Although there are fewer females dispersing than males, neither show significant isolation by distance when using pairwise relatedness values (Figure 3). In fact, when comparing all pairwise genetic distances, none was significant (P>0.05). This lack of differentiation over large distances (∼230 km) typifies the high levels of dispersal within these populations.
When examining genetic differentiation on a continental scale, there was strong evidence of genetic isolation between NSW and QLD regions. This result is clearly illustrated using both mtDNA and microsatellite data. For example, hierarchical analysis of mtDNA variance using AMOVA (Excoffier et al, 1992) partitioned a significant proportion of genetic variation (49%) between the two areas. In addition, average sequence divergence between the two regions was high (4.5%), with haplotypes being phylogenetically distinct (see Phylogeography). Furthermore, the multilocus clustering method of Pritchard et al (2000) indicated that NSW and QLD were almost certainly (P=0.999) two separate populations. These data suggest that both regions are, or have been, isolated for a significant period of time, at least with respect to maternal gene flow. However, this does not exclude the possibility of admixture or gene flow occurring at a contact zone between our sampled NSW and QLD populations. Consequently, further sampling is needed to clarify the extent of this genetic subdivision.
Phylogeography
Subspecies are largely regional variants of a species, and hence can be related to an evolutionary significant unit (ESU) (Moritz, 1994). Under this definition, the lack of genetic distinction (reciprocal monophyly) between M. g. giganteus and M. g. tasmaniensis does not reflect current taxonomic classification. Although morphological differences exist (Kirsch and Poole, 1972), they probably reflect clinal adaptation to environmental differences and/or stochastic effects associated with small founder populations, rather than taxonomic separation. Consequently, our data indicate that only one ESU exists for M. giganteus, and that the use of the present taxonomic subdivision within Australia should be reassessed.
Within mainland Australia, there is clear phylogeographic distinction between northern and southern populations. Although there is no reciprocal monophyly between these groups, the results suggest significant isolation for an extensive period of time. Owing to the lack of samples connecting these two areas, the cause of these differences is unknown, although there are relatively few physical barriers to dispersal. Within each of these groups, however, variant haplotypes are spread throughout all sampled populations (especially in southern Australia). This pattern implies that regional populations have been connected by large amounts of historical and/or current mtDNA geneflow. The limited divergence among northern haplotypes compared to southern, suggests a relatively recent colonization of QLD by M. giganteus. As two mtDNA lineages from southern Australian are basal to the QLD lineage, it seems likely that a small number of southern M. giganteus originally colonized northeastern Australia and that control region variation has since been generated via mutation.
Implications for management
Management of kangaroo populations whether for conservation, or as a renewable resource, or for pest control, requires effective population monitoring and understanding of the biology of the species. The findings of this study challenge previous ecological studies by providing a perspective on parameters such as historical and contemporary population structure and dispersal, difficult to obtain through direct observations. These results will aid in the future management of this species, by providing knowledge of the geographical scale at which M. giganteus populations are genetically distinct. Several important observations emerge from our findings. First, patterns of weak population differentiation within NSW are almost certainly a consequence of high gene flow among these geographically distinct populations (up to 230 km apart). Consequently, management regimes will need to focus on a regional scale rather than a local scale to effectively manage these populations. In addition, females should be targeted, as these appear to have higher site fidelity and ultimately will preferentially control the rate of net recruitment within specific populations. Second, the lack of genetic differentiation within regions suggests that gene flow may over-ride local selection. This has implications for translocation of animals and/or reintroductions. Currently, in the interests of genetic integrity of kangaroo populations, NSW National Park and Wildlife Service policy states that the release of large macropods is constrained within 50 km of the encounter point (Hardy, 2000). Obviously, this geographical range may be increased regarding M. giganteus given the lack of substantial genetic differentiation among NSW populations. Thirdly, the current taxonomic status of mainland and Tasmanian M. giganteus populations should be revised to reflect separate populations, rather than separate subspecies. Lastly, the extent of phylogenetic separation between southern (NSW/VIC) and northern (QLD) M. giganteus populations and the apparent reduction in mtDNA diversity within northern populations are sufficient for these regions to be recognized as separate entities for management regimes on a continental scale.
References
Baker CS, Slade RW, Bannister JL, Abernethy RB, Weinrich MT, Lien J et al (1994). Hierarchical structure of mitochondrial DNA gene flow among humpback whales Megaptera novaeangliae, world-wide. Mol Ecol 3: 313–327.
Bowyer JC, Newell GR, Eldridge MDB (2002). Genetic effects of habitat contraction on Lumholtz's tree-kangaroo (Dendrolagus lumholtzi) in the Australian wet tropics. Conserv Genet 3: 61–69.
Caughley G, Sinclair ARE (1994). Wildife Ecology and Management. Blackwell Scientific Publishers: London.
Clegg SM, Hale P, Moritz C (1998). Molecular population genetics of the red kangaroo (Macropus rufus): mtDNA variation. Mol Ecol 7: 679–686.
Coulson G (1998). Management of overabundant macropods - are there conservation benefits? In: Austin A, Cowan P. (eds) Managing Marsupial Abundance for Conservation Benefits. Issues in Marsupial Conservation and Management, Occasional Papers of the Marsupial CRC No. 1, Sydney pp 37–48.
Coulson G, Alviano P, Ramp P, Way S (1999). The kangaroos of Yan Yean: history of a problem population. Proc R Soc Vic 111: 121–130.
Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Firestone KB, Elphinstone MS, Sherwin WB, Houlden BA (1999). Phylogeographical population structure of tiger quolls Dasyurus maculatus (Dasyuridae: Marsupialia), an endangered carnivorous marsupial. Mol Ecol 8: 1613–1625.
Frankham R (1996). Relationship of genetic variation to population size in wildlife. Conserv Biol 10: 1500–1508.
Frith HJ, Calaby JH (1969). Kangaroos. Cheshire Publishing: Melbourne.
Fumagalli L, Pope LC, Taberlet P, Moritz C (1997). Versatile primers for the amplification of the mitochondrial DNA control region in marsupials. Mol Ecol 6: 1199–1201.
Goodnight KF, Queller DC, Poznansky T (1998). KINSHIP (version 1.2). Good-night software. Available at http://www.bioc.rice.edu/~kfg/Gsoft.html.
Goudet J (2000). FSTAT (version 2.9.1) A program to estimate and test gene diversities and fixation indices. Available at http://www.unil.ch/izea/softwares/fstat.html.
Hardy J (2000). Wildlife rehabilitation (lecture notes for training course by NPWS). Wildlife licensing, NSW National Parks & Wildlife Service.
Hill GJE, Barnes A, Wilson GR (1988). The use of wheat crops by grey kangaroos, Macropus giganteus, in southern Queensland. Aust Wildl Res 15: 111–117.
Jaremovic RV, Croft DB (1991). Social organization of the eastern grey kangaroo (Macropodidae, Marsupialia) in southeastern New South Wales. I. Groups and group home ranges. Mammalia 55: 169–185.
Jarman PJ, Taylor RJ (1983). Ranging of eastern grey kangaroos and wallaroos on a New England pastoral property. Aust. Wildl. Res, 10: 33–38.
Johnson CN (1989). Dispersal and philopatry in the macropodoids. In: Grigg G, Jarman PJ, Hume I (eds) Kangaroos, Wallabies and Rat-Kangaroos, Surrey Beaty & Sons: Sydney. pp 593–601.
Kaufmann JH (1975). Field observations of the social behaviour of the eastern grey kangaroo, Macropus giganteus. Anim Behav 23: 214–221.
Kimura M (1980). A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120.
Kirsch JAW, Poole WE (1972). Taxonomy and distribution of the grey kangaroo, Macropus giganteus Shaw and Macropus fuliginosus (Desmarest) and their sub-species (Marsupialia: Macropodidae). Aust J Zool 20: 315–339.
Kumar S, Tamura K, Jakobsen I, Nei M (2001). MEGA2: Molecular Evolutionary Genetics Analysis software. Available at http://www.megasoftware.net/.
Mantel N (1967). The detection of disease clustering and a generalised regression approach. Cancer Res 27: 209–220.
McElroy D, Moran P, Bermingham E, Kornfield I (1991). REAP: the restriction enzyme analysis package version 4.0. J Hered 83: 157–158.
Moritz C (1994). Defining ‘Evolutionary Significant Units’ for conservation. Trends Ecol Evol 9: 373–375.
Moritz C, Lavery S (1996). Molecular ecology: contributions from molecular genetics to population ecology. In: Floyd RB, Sheppard AW, De Barro PJ (eds) Frontiers of Population Ecology, CSIRO Publishing: Melbourne. pp 433–450.
Poole WE (1982). Macropus giganteus. Mamm Species 187: 1–8.
Poole WE (1995). Eastern grey kangaroo, Macropus giganteus (Shaw, 1790). In: Strahan R (ed) The Mammals of Australia, Reed Books: Sydney.
Pope LC, Sharp A, Moritz C (1996) Population structure of the yellow-footed rock-wallaby Petrogale xanthopus (Gray, 1854) inferred from mtDNA sequences and microsatellite loci. Mol Ecol 5: 629–640.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Raymond M, Rousset F (1995). GENEPOP (version 1.2) Population Genetics Software for Exact Tests and Ecumenicism. J Hered 86: 248–249.
Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Schneider S, Kueffer JM, Roessli D, Excoffier L (1997). ARLEQUIN (version 2.0). A software for population genetic data analysis. Available at http://anthro.unige.ch/software/arlequin/.
Schneider S, Excoffier L (1999). Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152: 1079–1089.
Simon JL (1999). RESAMPLING STATS (version 5.0.2). Resampling Stats Inc. Available at http://www.resample.com.
Slatkin M (1993). Isolation by distance in equilibrium and non-equilibrium population. Evolution 47: 264–279.
Spencer PB (1996). Coping with a naturally fragmented environment: a genetic and ecological study of the Allied rock-wallaby, Petrogale assimilis. PhD Thesis, James Cook University, Queensland.
Spencer PB, Odorico DM, Jones SJ, Marsh HD, Miller DJ (1995). Highly variable microsatellites in isolated colonies of the rock-wallaby (Petrogale assimilis). Mol Ecol 4: 523–525.
Sunnucks P (2000). Efficient genetic markers for population biology. Trends Ecol Evol 15: 199–203.
Sunnucks P, Hales DF (1996). Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol Biol Evol 13: 510–524.
Sunnucks P, Wilson AC, Beheregaray LB, Zenger KR, French J, Taylor AC (2000). SSCP is not so difficult: the application and utility of single-stranded conformation polymorphism in evolutionary biology and molecular ecology. Mol Ecol 9: 1699–1710.
Swofford DL (2000). PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) version 4. Sinauer Associates: Sunderland, MA.
Swofford DL, Selander RB (1981) BIOSYS-1: a FORTRAN program for the comprehensive analysis of electrophorestic data in population genetics and systematics. J Hered 72: 281–283.
Taylor AC, Cooper DW (1998) A set of tammar wallaby (Macropus eugenii) microsatellites tested for genetic linkage. Mol Ecol 7: 925–926.
Taylor AC, Sherwin WB, Wayne RK (1994). Genetic variation of microsatellite loci in a bottlenecked species: the northern hairy-nosed wombat Lasiorhinus krefftii. Mol Ecol 3: 277–290.
Wakeley J (1993). Substitution rate variation among sites in hypervariable region 1 of human mitochondrial DNA. J Mol Evol 37: 613–623.
Wright S (1951). The genetic structure of populations. Ann Eugen 15: 323–354.
Zenger KR, Cooper DW (2001a). Characterization of 14 macropod microsatellite genetic markers. Anim Genet 32: 166–167.
Zenger KR, Cooper DW (2001b). A set of highly polymorphic microsatellite markers developed for the eastern grey kangaroo (Macropus giganteus). Mol Ecol Notes 1: 98–100.
Acknowledgements
We thank the many people who helped collect samples, especially Tony English, Graeme Coulson, Peter Johnson and Steve Cronin. We are also grateful to Adam Stow for expert analytical advice and Professor Trevor Tansley, Dean of Graduate Students, Macquarie University and Professor D Yerbury, Vice Chancellor, Macquarie University for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zenger, K., Eldridge, M. & Cooper, D. Intraspecific variation, sex-biased dispersal and phylogeography of the eastern grey kangaroo (Macropus giganteus). Heredity 91, 153–162 (2003). https://doi.org/10.1038/sj.hdy.6800293
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800293
Keywords
This article is cited by
-
Demographic drivers of Norway rat populations from urban slums in Brazil
Urban Ecosystems (2021)
-
Redescription of Rugopharynx australis (Mönnig, 1926) and the description of R. moennigi n. sp. (Nematoda: Strongyloidea) from kangaroos (Marsupialia: Macropodidae) in Australia
Systematic Parasitology (2021)
-
The genetic relatedness of a peri-urban population of eastern grey kangaroos
BMC Research Notes (2018)
-
Molecular and microscopic characterization of a novel Eastern grey kangaroopox virus genome directly from a clinical sample
Scientific Reports (2017)
-
Sex-linked and autosomal microsatellites provide new insights into island populations of the tammar wallaby
Heredity (2014)