Abstract
Heritable variation in fitness is the fuel of adaptive evolution, and sex can generate new adaptive combinations of alleles. If the generation of beneficial combinations drives the evolution of recombination, then the level of recombination should result in changes in the response to selection. Three types of lines of Drosophila melanogaster varying in their level of genetic recombination were selected over 38 generations for geotaxis. The within-chromosome recombination level of these lines was controlled for 60% of the genome: chromosome X and chromosome II. The full recombination lines had normal, unmanipulated levels of recombination on these two chromosomes. Conversely, nonrecombination lines had recombination effectively eliminated within the X and second chromosomes. Finally, partial recombination lines had the effective rate of within-chromosome recombination lowered to 10% of natural levels for these two chromosomes. The rate of response to selection was measured for continuous negative geotaxis and for a fluctuating environment (alternating selection for negative and positive geotaxis). All selected Drosophila lines responded to selection and approximately 36% of the response to selection was because of the X and second chromosomes. However, recombination did not accelerate adaptation during either directional or fluctuating selection for geotaxis.
Similar content being viewed by others
Introduction
Although the evolutionary value of sex is the promotion of recombination, understanding the forces maintaining sex and recombination remains one of the most difficult problems in evolutionary theory (Barton and Charlesworth, 1998). Several theoretical explanations have already been formulated, which fall into two broad categories (Otto and Barton, 1997). Recombination may have been selected because it plays a critical physiological role in the life of the cell (physiological hypotheses, see Otto and Barton, 1997) and/or because it promotes genetic variability in the face of selection (generative hypotheses, see Otto and Barton, 1997). In this latter case, recombination can be favored if it generates under-represented, fit genotypes or produces novel, beneficial allelic combinations.
During directional selection, increased recombination may be favored even in the absence of epistatic interactions. This may happen when recombination is selected on its ability to protect beneficial alleles from stochastic loss (Fisher, 1930; Muller, 1932). Indeed in a finite population, a new beneficial mutation is more likely to fix if it is unlinked to other selected loci; the higher the recombination, the higher the probability of fixation (Hill and Robertson, 1966; Barton, 1995a). Otto and Barton (1997) showed that a modifier allele that increases recombination can be selected for because it increases the fixation of beneficial alleles, even if there are no epistatic interactions. Hence during directional selection, a modifier allele that increases recombination increases the chance that new beneficial alleles will fix and, as these alleles rise to fixation, they carry the modifier allele along with them by hitchhiking.
If the evolution of recombination is driven by the fate of beneficial mutations (with linkage disequilibrium generated either by negative epistasis or stochastically) rather than by the removal of deleterious mutations, directional selection for fitness traits (eg traits involved in adaptation to unfavorable conditions) may result in an increase in the level of recombination. This has been verified in a number of empirical studies (eg Harinarayana and Murty, 1971; Flexon and Rodell, 1982; Zhuchenko et al, 1985; Burt and Bell, 1987; Wolf et al, 1987; Korol and Iliadi, 1994). The reverse prediction – that is that manipulating the level of recombination should result in changes in the response to selection – has received some attention to date, with contradictory results (Carson, 1958; McPhee and Robertson, 1970; Markow, 1975; Malmberg, 1977; Thompson, 1977; Zeyl and Bell, 1997; Rice and Chippendale, 2001).
In each of these latter experiments, however, the change in recombination rate is confounded with the mechanism used to reduce recombination. This problem is avoidable in Drosophila, where the males naturally have extremely low rates of recombination. We take advantage of this fact by doing selection only on males with wild-type karyotypes, but with the suppression of recombination in some females using balancer chromosomes. In this paper, we use Drosophila melanogaster to determine: (i) whether variation in the level of recombination induces changes in the response to artificial selection for positive and negative geotaxis behavior, (ii) whether a low level of recombination is sufficient to produce a significant increase in response to selection compared to the absence of crossingover and (iii) if fluctuation in the direction of selection alters the response to selection for geotaxis.
Materials and methods
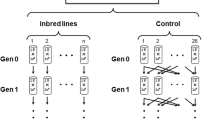
The rate of within-chromosome recombination was controlled by the use of balancer chromosomes in replicate lines of D. melanogaster, undergoing different types of selection for geotaxis. There were four types of lines: nonselection controls and three kinds of selected lines differing in recombination:
-
control lines (C) had natural levels of recombination but no artificial selection;
-
full recombination lines (FR) likewise had normal, unmanipulated levels of recombination;
-
nonrecombination lines (NR) had recombination effectively eliminated for the X and second chromosomes; and
-
partial recombination lines (PR) had the effective rate of within-chromosome recombination lowered to 10% of natural levels for the X and second chromosomes.
In order to control the rates of recombination, two very useful aspects of Drosophila biology and technology were put to use. First, males of D. melanogaster have very low levels of recombination naturally. Thus to control recombination rates, we need only control the rate of recombination in females. Second, balancer chromosomes are available, which effectively prevent recombination in heterozygotes. These balancers are also genetically marked so that their carriers can be distinguished from wild-type homozygotes. Thus, the recombination rate of a population can be controlled by the proportion of chromosomes that are paired with balancer chromosomes in females.
Two different balancer chromosomes were used for this purpose. Fm7 and CyO are balancer chromosomes for the X and second chromosomes, respectively. These two were used to stop recombination in individuals that carried them; the relative numbers of individuals carrying Fm7 and CyO depended on the experimental treatment. Recombination was not experimentally manipulated for the third or fourth chromosomes; the third chromosome carries about 40% of the genetic material in D. melanogaster, while the fourth chromosome has less than 1%.
These lines were exposed to varying types of selection. The controls were not subject to artificial selection, but were measured for each generation and otherwise their husbandry was similar to the selected lines. All other lines were subjected for 15 generations to select for negative geotaxis (ie to go up), after which each line was divided into two. For each of these new pairs of lines, one line continued to be selected for negative geotaxis for 15 more generations (called up lines) and the other was subjected to selection for positive geotaxis for 15 generations followed by eight further generations of selection for negative geotaxis (the Fluctuating lines). More details will be given in the following sections.
Derivation of the lines
All flies were maintained in 75 × 25 mm standard food vials with 7 ml of corn meal/agar medium at 25±1°C with a 12:12 light:dark cycle. All handling was performed at room temperature using CO2 anesthesia.
All lines were derived from the Dahomey laboratory population. Before the founding of the lines, a new stock population was created that had the genotype Fm7/+; CyO/+; +/+, where the +'s indicate multiple wild-type chromosomes derived from the Dahomey population. This stock was created with a large number of wild-type founders and therefore contains nearly all of the genetic variation from the original stock.
Five control lines were each started from 50 virgin females and 50 randomly chosen males from the Dahomey population. Five replicates of each of the other types of line were also created; for each of the selected lines (FR, PR, and NR) the males used to found the lines were the 50 which showed the most negative geotaxis out of 500 randomly chosen males from the Dahomey stock (see below for selection details). A separate 500 flies was measured for each of the 15 selected lines. The females founding the selected lines were taken in combinations from the Dahomey stock and the balancer/Dahomey stock such that there were a total of 100 wild-type chromosomes from the Dahomey population, but where the level of recombination would match the description of the population. Thus, for the FR lines the founding females for each were a random sample of 50 individuals from the Dahomey population. For the NR lines, 100 females from the balancer/Dahomey population, where each female is heterozygous for both balancers and therefore carries only a single copy of each wild-type chromosome, were used to found each line. For the PR lines, 90 females from the balancer/Dahomey line and five females from the Dahomey stock were used.
After the first generation, each line was perpetuated by random mating between the selected males and virgin females taken in proportion to the desired recombination level. In the first 15 generations, the numbers of males and females were taken from the previous generation of the same line and followed the same numbers as at founding: 50 wild-type males (selected from 500, except in the controls) and females chosen for the number of genotypes as and when they were founded. In the PR lines, the preselection males in each generation were taken in 90:10 proportions from the nonrecombination: recombination females in the previous generation. After the 15th generation, the lines were subdivided into different selection treatments (Up and Fluctuating); at this point the population sizes were halved: 25 males were selected from 250, and there were a total of 50 wild-type chromosomes in mating females each generation. For the PR lines, the ratio of balanced to wild-type females then changed slightly: two females without balancers and 46 heterozygotes for the balancers were used in these lines in the second half of the experiment.
Selection
Measurement of the geotaxis preferences of individual flies was performed in vertical mazes similar to those described in Hirsch (1959). The mazes were constructed of plastic tubing of 8 mm interior diameter, joined by plastic cross-junctions (see Figure 1). In each junction a funnel made of a truncated pipette tip was wedged; these acted as unidirectional filters, so that flies did not go backwards in the maze. Male flies were introduced into the beginning of the maze, and then each fly faced 10 up/down choices before exiting the maze into collection vials. The vial in which the flies started was without food, but the collection vials at the end of the maze contained standard yeasted food. The top collection vial (vial #1) was filled with flies that made 10 separate ‘up’ choices, against gravity. Conversely, the bottom vial (vial #10) was filled with flies that made 10 separate ‘down’ choices (see Figure 1). A fly that went up nine out of 10 times but chose to go down once would wind up in vial #2, etc.
Each maze was mounted to a vertical board kept in a light-tight box to ensure complete darkness in order to avoid phototaxis. The temperature was kept constant at 25°C in the room in which the box was stored. When a replicate line was run through the maze, all four types of line or subline were run at the same time in other mazes. Each line was run through the maze for 24 h, then the collection vials were removed for counting and selection.
For the first 15 generations of the experiment, 500 males were run through a maze for each experimental line, from which 50 were chosen in accordance with the type of selection. That is, for the selected lines, the 50 males that went highest in the maze were kept for breeding the following generation; for the control lines 50 males were randomly chosen from all those found in any collection vial. For the 16th and following generations, 250 males were run though the maze for each line, and 25 were selected to form the next generation. In lines where selection was reversed, the males were taken from the lowest collection vials.
Note that in all generations and all lines, only wild-type males were used in the maze; the fact that male Drosophila do not have recombination means that it is unnecessary to control their recombination rate. Thus, no males with balancer chromosomes were used in the experiment, so selection was not influenced by differences in either mean geotaxis rates in the balancers versus wild-type flies, nor do any differences in genetic variance in the base stocks matter.
Effect of the X and second chromosomes
The experiment was designed to estimate the influence of recombination on the first and second chromosomes. Recombination was not controlled on the third chromosome. We therefore wished to know the extent to which the response to selection was because of the two manipulated chromosomes. At generations 18, 30, and 38, each line was crossed in a pedigree, which allowed the third chromosome to be replaced by a marked standard chromosome, while keeping the first two chromosomes polymorphic for chromosomes taken from the lines. In total, 500 males from each of these swapped lines were then measured in the geotaxis maze.
The third chromosome swap was performed in the following way. We created a stock of flies that had the genotype Fm7/+; CyO/Bl; e/e. Bl is a dominant bristle mutation, and ebony (e) is a recessive body color mutation. The third chromosomes carrying e in this stock had low genetic variance, having all been derived from two ancestral chromosomes. Wild-type females from each line were crossed to a male from this ebony stock. Sons from this cross were crossed to females from the ebony stock. This cross produced (among other discarded genotypes) individuals that were homozygous for the ebony chromosome and that had X and second chromosomes from the experimental line balanced by Fm7 (or a Y chromosome) and CyO. Males and females from these individuals were crossed and the wild-type male offspring were saved for measurement in the geotaxis maze. These males had all of their X and second chromosomes from the experimental line and both third chromosomes from the ebony stock. The Y chromosome also came from the ebony stock, and the small fourth chromosome was uncontrolled.
Results
Response to selection
All of the treatments significantly responded to selection (Figure 2). The P-values for this response are given by the heritability analysis below. The mean phenotype of the selected lines at the end of each phase of selection was sufficiently close to the maximum (ie the limits of the maze) that continued selection would likely have been relatively ineffective.
The mean response to selection in the different selected lines over generation number. The geotaxis phenotype attributed to each fly was the number of the vial into which it emerged from the maze (see Figure 1). Thus flies arriving in the upper vial (exhibiting negative geotaxis) had a score of 1, and flies in the lowest vial (exhibiting positive geotaxis behavior) had a score of 10. For this figure, the mean geotaxis score calculated over the five replicates was subtracted from the mean of the controls. FR up, PR up, and NR up lines were selected for negative geotaxis over 30 generations. At generation 15, each selected line was used to generate new lines referred to as FR fluctuating, PR fluctuating, and NR fluctuating. These fluctuating lines were selected for positive geotaxis for 15 generations followed by selection for negative geotaxis for eight more generations.
However, much of this response to selection was because of the effects of the third chromosome, for which recombination was not experimentally controlled. This is shown by the fact that in the lines in which the third chromosome was genetically crossed out, the change in mean phenotype over time was much reduced (see Table 1). Nevertheless, after 30 generations of selection, the lines consistently selected to go up had more negative geotaxis (Wilcoxon sign-rank tests for difference from control: FR, P=0.047; PR, P=0.028; NR, P=0.076) even when the third chromosome was swapped, but there was no significant heterogeneity in the response to selection of the different recombination types (P=0.83, Kruskal–Wallis test). Further evidence that the X and second chromosomes were involved in some of the response to selection comes from the divergence of the paired lines between generations 15 and 30, when each line was divided into sublines. Pooling over all selected lines, the difference at generation 30 of the up and down sublines with their third chromosomes replaced was significant (P=0.005 by one-tailed paired t-test). Given the low number of lines for each type, the differences were not significant for all of the recombination types considered individually (FR, P=0.13; PR, P=0.012; NR, P=0.18). The mean difference in the chromosome-replaced selected sublines at generation 30 was 1.4, while the mean difference in the lines including the effects of the selected third chromosomes was 3.94. This implies that approximately 36% of the response to selection was because of the X and second chromosomes, the rest because of chromosome III.
Heritabilities
The most powerful way of determining whether recombination rate affects the response to selection is to compare the heritabilities (h2) of the different lines. To account for fluctuations in the experimental conditions over generations, for each generation the mean value of the control lines in that generation was subtracted from the values of each individual. These residuals were used for all analyses. For each subline, the heritability was calculated for each seven or eight generations, by regressing the cumulative response to selection by generation on the cumulative selection differential (Walsh and Lynch, in preparation). For the first set of generations, this regression was forced through zero at the beginning, but for subsequent generations the starting means are not known with certainty, so an intercept was fitted as well. The slope of the regression of cumulative response to selection by generation on the cumulative selection differential gives an estimate of the heritability.
The mean heritabilities of each type are given in Table 2. For all recombination treatments, there was evidence of significant heritability during most periods of the experiment. However, there was little or no difference between the recombination treatments in the level of heritability. For the sublines consistently selected to go up, there were no significant differences between the heritabilities of the different recombination types, for any of the time periods (by Kruskal–Wallis tests, the P-values were 0.28, 0.54, 0.88, and 0.68 for the four time periods indicated in Table 2, respectively). With fluctuating selection, there was a significant difference in the last time period only (P-values 0.14, 0.21, 0.014, respectively). This last result is not significant (P=0.1) when corrected for the seven multiple comparisons by the Bonferroni method. It is difficult to interpret the result for this last time period, as the partial recombination lines have the highest heritability, with no difference between the full and zero recombination lines. There is very little evidence from these data that there is any difference in realized heritability among treatments with very different levels of recombination.
The cumulative P-value for the test for differences across treatments across all these time divisions can be approximated by using the sum of χ2 values across all the Kruskal–Wallis tests. This is only an approximation, because the different tests are not independent (they use the same lines), but it should be biased towards finding differences where none exist. Even with this more powerful test, there is no evidence for differences between recombination treatments in their cumulative response to selection (P=0.12).
Discussion
Our results show that Drosophila responds to selection for geotaxis, but within-chromosome recombination did not accelerate response to selection. A more substantial response was expected in the lines undergoing fluctuating selection because this regime of selection is thought to be favorable for the evolution of high rates of recombination (Sasaki and Iwasa, 1987; Barton, 1995b). However, when alternating selection for positive and negative geotaxis, we did not find substantial differences among recombinant, partially recombinant, or nonrecombinant populations.
Such a negative result has previously been reported by Zeyl and Bell (1997), studying the effect of sex on mean fitness in experimental populations of the budding yeast Saccharomyces cerevisiae. At the end of their experiment, the sexual populations appeared no better adapted to galactose than the asexual populations. Their explanation was the presence of a single locus with major effect. Indeed, recombination is expected to be advantageous only if multiple mutations were responsible for adaptation. In the case of geotaxis in Drosophila, little is known about the genetic basis, except that multiple chromosomes can contribute to the response to selection in this trait (see below).
Other experiments have found an effect of recombination on the response to selection, for example, McPhee and Robertson (1970). In their experimental populations of D. melanogaster, suppression of recombination on chromosomes II and III slowed the response to artificial selection on sternopleural bristle numbers. However, this experiment has a small flaw: the effect of recombination is confounded by the presence of balancer chromosomes in some of the selected individuals. The balancer stocks may have had different levels of genetic variation than the wild-type stocks, which could explain the difference in response to selection that McPhee and Robertson observed. (The experiments of Thompson (1977), Carson (1958), and Markow (1975) share this problem.)
Other experiments have suggested an advantage of recombination. The evolution of polygenic proflavine resistance in T4 viruses showed that populations with more recombination responded faster to selection, a difference that was consistent under three somewhat different conditions (Malmberg, 1977). Rice and Chippendale (2001) found a much greater response to selection at a single locus with recombination than without.
In D. melanogaster, geotaxis is one of the genetically best-studied behavioral traits. Erlenmeyer-Kimling and Hirsch (1961) analyzed populations selected for positive and negative geotaxis, and showed that this character was controlled by additive polygenic factors located on chromosomes X, II, and III. The polygenic nature of geotaxis was confirmed by Hirsch and Erlenmeyer-Kimling (1962), Hostetter and Hirsch (1967), and Walton (1968). Hirsch and Erlenmeyer-Kimling (1962) found that, in an unselected wild-type population, chromosomes X and II contributed to positive geotaxis, while chromosome III contributed to negative geotaxis. In the strain that had been selected for positive geotaxis, all three major chromosomes contributed to the determination of the sign of geotaxis, while in the strain selected for negative geotaxis, the positive effect of X and II was strongly reduced although still present, while the negative effect of III was considerably increased.
This genetic analysis is consistent with the results obtained in a study that looked at the effects of selection on recombination rates. Korol and Iliadi (1994) selected both positively and negatively for geotaxis in D. melanogaster and observed substantial increases in recombination. Across all regions studied, the map length rose from 217.0 cM in the control line to 295.9 and 283.4 cM in the lines selected for negative and positive geotaxis over a period of 50 generations, respectively. All of the three large chromosomes showed significant increases in map length in the positively selected lines and two (chromosomes X and III) showed significant increases in negatively selected lines. Our results indicate that the response to negative geotaxis was mostly – although not completely – because of genes located on the third chromosome: in the lines in which the third chromosome was genetically replaced, the change in mean phenotype over time was much reduced, but still significant.
Our results show that reducing the level of within-chromosome recombination to effectively zero for 60% of the genome has negligible consequences for the rate of response to selection, either with directional or fluctuating selection. In part, this is because of the fact that geotaxis is affected largely by genetic variation on the uncontrolled third chromosome, but even when that chromosome is experimentally removed, there is no difference in the response to selection as a function of recombination rate. Recombination does not necessarily change the rate of adaptation.
There is, however, a caveat to this conclusion. Like all negative results, we must ask whether there is sufficient power to make a broad claim. The response to selection was very variable within experimental treatments, and this reduces the power to discriminate true differences among treatments. In fact, a power analysis shows that, for a comparison of heritabilities among treatments (averaged over each time period), a 20% change in h2 would have about a 24% chance of being detected with this experimental design, while a 50% change in h2 would have been detected about 80% of the time with this design. Clearly, there is a good chance of missing real differences, but improving the power would have been difficult, as the amount of labor involved in this experiment was enormous. This study should be considered in the context of other similar studies, if such a case be carried out the future. A key component of future studies, however, should be the fact that the individuals under selection do not differ in their marker genotypes or in the stocks from which they are derived. A great deal of the labor associated with this experiment has been to assure that this basic control has been observed. This distinguishes this study from, say, that of McPhee and Robertson (1970).
Another issue is that while we controlled for the level of recombination within two of the chromosomes, we did not control for segregation among chromosomes. If the key element controlling the evolution of recombination is the probability that two randomly chosen loci are separated during meiosis, then segregation is more important than within-chromosome recombination. We reduced this probability by 20% in our NR lines. Most analyses of recombination, though, are focused on understanding the evolution of recombination within chromosomes and, in particular, over short genetic distances. Our results give no support for recombination rates within chromosomes evolving in response to directional selection on a quantitative trait.
Two different kinds of explanations for the evolution of recombination have been proposed (Michod and Levin, 1988). One is that the genes responsible for recombination are needed to repair DNA and ensure proper chromosomal segregation. Recombination genes may also produce new genetic variability that is important in evolution, but this could be seen as an unselected consequence, rather than the cause of the evolution of recombination. The alternative view is that the role of recombination genes in producing new gene combinations by crossingover has itself been an important selective force in their evolution. The present experiment does not support this generative hypothesis. More experimental manipulations of selection and recombination will be necessary to resolve the causes and effects of this important trait.
References
Barton NH (1995a). Linkage and the limits to natural selection. Genetics 140: 821–841.
Barton NH (1995b). A general model for the evolution of recombination. Genet Res 65: 123–144.
Barton NH, Charlesworth B (1998). Why sex and recombination? Science 281: 1986–1990.
Burt A, Bell G (1987). Mammalian chiasma frequencies as a test of two theories of recombination. Nature 326: 803–805.
Carson HL (1958). Response to selection under different conditions of recombination in Drosophila. Cold Spring Harbor Symp Quant Biol 23: 291–306.
Erlenmeyer-Kimling L, Hirsch J (1961). Measurement of the relations between chromosomes and behavior. Science 134: 1068–1069.
Fisher RA (1930) The Genetical Theory of Natural Selection. Oxford University Press: Oxford.
Flexon PB, Rodell CF (1982). Genetic recombination and directional selection for DDT resistance in Drosophila melanogaster. Nature 298: 672–674.
Harinarayana G, Murty BR (1970). Cytological regulation of recombination in Pennisetum and Brassica. Cytologia 36: 435–448.
Hill WG, Robertson A (1966). The effect of linkage on the limits to artificial selection. Genet Res 8: 269–294.
Hirsch J (1959). Studies in experimental behavior genetics: II. Individual differences in geotaxis as a function of chromosome variations in synthesized Drosophila populations. J Comp Physiol Psychol 52: 304–308.
Hirsch J, Erlenmeyer-Kimling L (1962). Studies in experimental behavior genetics: IV. Chromosome analyses for geotaxis. J Comp Physiol Psychol 55: 732–739.
Hostetter RC, Hirsch J (1967). Genetic analysis of geotaxis in Drosophila melanogaster: complementation between forward and reverse selection lines. J Comp Physiol Psychol 63: 66–70.
Korol AB, Iliadi KG (1994). Increased recombination frequencies resulting from directional selection for geotaxis in Drosophila. Heredity 72: 64–68.
Malmberg RL (1977). The evolution of epistasis and the advantage of recombination in populations of bacteriophage T4. Genetics 86: 607–621.
Markow TA (1975). A genetic analysis of phototactic behavior in Drosophila melanogaster I. Selection in the presence of inversions. Genetics 79: 537–534.
McPhee CP, Robertson A (1970). The effect of suppressing crossing-over on the response to selection in Drosophila melanogaster. Genet Res 16: 1–16.
Michod RE, Levin BR (1988). The Evolution of Sex: An Examination of Current Ideas. Sinauer: Sunderland.
Muller HJ (1932). Some genetic aspects of sex. Am Nat 66: 118–138.
Otto SP, Barton NH (1997). The evolution of recombination: removing the limits to natural selection. Genetics 147: 879–906.
Rice WR, Chippendale AK (2001). Sexual recombination and the power of natural selection. Science 294: 555–559.
Sasaki A, Iwasa Y (1987). Optimal recombination rate in fluctuating environments. Genetics 115: 377–388.
Thompson V (1977). Recombination and response to selection in Drosophila melanogaster. Genetics 85: 125–140.
Walsh B, Lynch M . Evolution and Selection of Quantitative Traits. MS in preparation.
Walton PD (1968). The genetics of geotaxis in Drosophila melanogaster. Can J Genet Cytol 10: 673–687.
Wolf HG, Wohrmann K, Tomiuk J (1987). Experimental evidence for the adaptive value of sexual reproduction. Genetica 72: 151–159.
Zeyl C, Bell G (1997). The advantage of sex in evolving yeast populations. Nature 388: 465–468.
Zhuchenko AA, Korol AB, Kotyukh LP (1985). Change of crossing-over frequency in Drosophila during selection for resistance to temperature fluctuations. Genetica 67: 73–78.
Acknowledgements
We thank S Otto, A Peters, and two anonymous reviewers for comments on the manuscript. Funding was provided by a grant from the Natural Science and Engineering Research Council (Canada).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bourguet, D., Gair, J., Mattice, M. et al. Genetic recombination and adaptation to fluctuating environments: selection for geotaxis in Drosophila melanogaster. Heredity 91, 78–84 (2003). https://doi.org/10.1038/sj.hdy.6800283
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800283
Keywords
This article is cited by
-
Y-chromosomes can constrain adaptive evolution via epistatic interactions with other chromosomes
BMC Evolutionary Biology (2018)
-
Experimental evolution of recombination and crossover interference in Drosophila caused by directional selection for stress-related traits
BMC Biology (2015)
-
The impact of recombination on short-term selection gain in plant breeding experiments
Theoretical and Applied Genetics (2013)
-
Natural Variation in Drosophila Stressed Locomotion Meets or Exceeds Variation Caused by Hsp70 Mutation: Analysis of Behavior and Performance
Behavior Genetics (2009)
-
An evolutionary view of human recombination
Nature Reviews Genetics (2007)