Abstract
Fine-scale structure of genetic diversity and gene flow were analysed in three Costa Rican populations of mahogany, Swietenia macrophylla. Population differentiation estimated using AFLPs and SSRs was low (38.3 and 24%) and only slightly higher than previous estimates for Central American populations based on RAPD variation (20%). Significant fine-scale spatial structure was found in all of the surveyed mahogany populations and is probably strongly influenced by the limited seed dispersal range of the species. Furthermore, a survey of progeny arrays from selected mother trees in two of the plots indicated that most pollinations involved proximate trees. These data indicate that very little gene flow, via either pollen or seed, is occurring between blocks of mahogany within a continuous or disturbed forest landscape. Thus, once diversity is removed from a forest population of mahogany, these data suggest that recovery would be difficult via seed or pollen dispersal, and provides an explanation for mahogany's apparent susceptibility to the pressures of logging. Evidence is reviewed from other studies of gene flow and seedling regeneration to discuss alternative extraction strategies that may maintain diversity or allow recovery of genetic resources.
Similar content being viewed by others
Introduction
To maintain natural populations as evolutionarily viable units able to adapt to changing conditions in the long term, genetic variation must exist. For selectively exploited tropical tree species, genetic erosion can become an important consideration and sustainable production can be threatened if genetically determined wood quality or pest resistance characters are undermined (Newton et al, 1996). A genetic resource management strategy for such species needs to be based on research data examining the extent of genetic differentiation within and between populations, and on understanding the processes maintaining this variation.
Mahogany, Swietenia macrophylla King (Meliaceae, Swietenioideae), is one of the most important timber species in the world (Rodan et al, 1992), and overexploitation and high rates of deforestation have led to it becoming the focus of increased conservation concern (Newton et al, 1996). Mahogany matures to a large, deciduous tree able to reach heights of 40 m, a diameter of over 2 m and may live for several centuries. It has a wide distribution, stretching from southern Mexico to the southern Amazon basin in Brazil (Pennington, 1981), and inhabits both wet and dry forests (rainfall typically 1000–2000 mm) and a range of soils up to 1400 m altitude (Whitmore, 1983). Although population densities are typically less than one individual per hectare (Barros et al, 1992), up to eight trees per hectare have also been recorded. Mahogany has unisexual, staminate and pistillate flowers (Styles and Khosla, 1976), which are insect-pollinated (bees, moths and thrips), and have a functional self-incompatibility system (Bawa et al, 1985; Mayhew and Newton, 1998), making it an obligate outbreeder. Although little information is available on phenology, flowering appears to be seasonal and simultaneous in most trees within a population. Leaf fall tends to occur at the onset of the dry season, leaving the mature seed capsules on the tree. Seed production from mahogany, in both natural and plantation trees, fluctuates considerably from year to year and may reflect variation in flowering phenology, or failure of pollination or fertilisation. The seeds are winged and dispersed by wind, but are relatively heavy, and tend to fall close to the maternal tree (32–36 m median distance). However, a distance of over 80 m was estimated for seed dispersal in a Bolivian population (Gullison et al, 1996).
In Meso-America deforestation rates are particularly rapid, where approximately 4% of forest is removed annually (Myers, 1989), but a large proportion of those forests that once contained mahogany has already been felled. A recent study using random amplified polymorphic DNA (RAPD) markers, found that 80% of the variation detected across Central American populations of S. macrophylla was maintained within populations (Gillies et al, 1999). This study also showed that a history of logging was correlated with a significant reduction in population diversity. However, reasons for the population level decline in genetic diversity following logging were not examined in detail. It is therefore a priority to examine the fine scale structure of genetic diversity and extent of gene flow within and between populations as a first step to understanding the potential impact of such landscape changes. Without this knowledge, it will be impossible to implement sustainable utilisation programmes for mahogany (Newton et al, 1999).
A range of powerful molecular genetic techniques, based on the polymerase chain reaction (PCR; Newton and Graham, 1994), are available for studying biodiversity, genetic structure and mating systems. In particular, the amplified fragment length polymorphism (AFLP) technique (Vos et al, 1995) targets random loci from across the plant genome, which are in general dominantly expressed. Owing to the large number of loci that can be screened using this technique, AFLPs are particularly applicable to studies of genomewide genetic diversity, structure and population differentiation. Another set of molecular markers, microsatellites (also called simple sequence repeats, or SSRs, Goldstein and Schlötterer, 1999), target individual loci of short tandem DNA repeats. The codominant and multiallelic nature of SSRs make them ideal for studying outcrossing rate and gene flow.
For the present study, three populations of S. macrophylla from northern Costa Rica were chosen, and were among those previously analysed by Gillies et al (1999) for RAPD variation. The aim of the investigation was to use AFLP analysis to examine the extent of fine-scale structure within all populations. In addition, primer pairs previously developed for microsatellite loci in S. humilis, but known to amplify in S. macrophylla (White and Powell, 1997), were used to assess outcrossing rate and potential gene flow in progeny cohorts from two of the populations. A combination of AFLP and SSR data will provide information on the population dynamics of S. macrophylla, and such investigations will contribute to the development of an integrated strategy for the sustainable resource management of S. macrophylla, which is an urgent priority.
Materials and methods
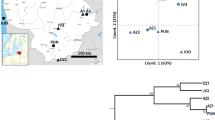
All populations selected for this study are located in northern Costa Rica (Figure 1). Pocosol is separated from the other two populations by an assemblage of mountains, the ‘Cordillera de Guanacaste’. Caño Negro and San Emilio are located in the wet region of Alajuela, east of the mountain range, while Pocosol is situated in the dry region of Guanacaste on the west coast. All populations have suffered some form of disturbance in their past. Caño Negro has been intensively logged, and most trees with superior form (ie tall, straight trunk) have been removed. The forest therefore only contains a low density of mahogany, but a few large trees remain in a disturbed forest landscape. San Emilio also has a history of logging. A proportion of trees of superior form have been removed, but the forest has been left to regenerate and forms a continuous canopy containing clusters of regenerating mahogany. The Pocosol study plot is located within a block of continuous forest with intact canopy, but is partially surrounded by grazing pasture. The Guanacasten name for the open woodland used for traditional grazing systems that characterise the Pocosol area are ‘sitios’, and it is likely that this area has been exploited over a long period of time (J Cornelius, personal communication). These historical observations are reflected in the density and average diameter at breast height (dbh) and height details in Table 1.
Leaf material was collected from 27 to 53 mapped mahogany trees from the three Costa Rican mahogany populations (Table 1). For molecular analysis, leaf samples were dried using silica gel (Weising et al, 1995). Seeds were collected from capsules found on the mother trees in Caño Negro (n=47) and Pocosol (n=20), the locations of which are shown in Figure 1. Seeds were germinated and progeny grown both at CATIE (Costa Rica) and CEH (Scotland). Total genomic DNA was extracted from dried (mature tree) or fresh (progeny) leaf material, using a modified CTAB extraction procedure (Gillies et al, 1999).
AFLP analysis followed the protocol of Vos et al (1995). Total genomic DNA was digested with the restriction enzymes EcoRI and MseI (Biolabs). Adaptors, consisting of the oligos CTCGTAGACTGCGTACC and AATTGGTACGCAGTCTAC for EcoRI and GACGATGAGTCCTGAG and TACTCAGGACTCAT for MseI, were ligated to the respective restriction sites. A selective preamplification PCR was carried out using an EcoRI primer (GACTGCGTACCAATTC) without additional nucleotides and an MseI primer (GATGAGTCCTGAGTAA) carrying two selective nucleotides (AT, AC, GC and CA). Final selective PCR reactions used EcoRI primers with two nucleotides and MseI primers with four nucleotides in the following combinations: Eco+GC/Mse+ATAG, Eco+GT/Mse+ACCC, Eco+CG/Mse+GCGA, Eco+CG/Mse+ACCG, Eco+GC/ Mse+CACG. All oligos and primers were obtained from Genset (Paris, France). The EcoRI primers were end labelled with [γ-33P] ATP and amplification products were visualised by polyacrylamide gel electrophoresis and exposure to Kodak X-ray film.
PCR amplification reactions using SSR primers contained: 10 mM of each of dATP, dTTP, dCTP and dGTP; 1 unit of Taq polymerase (Promega); 1/10th volume 10 × buffer (supplied with the enzyme); 200 pM microsatellite primers (MAC49, 54 and 58 from White and Powell, 1997); 1–10 ng of DNA; and sterile, deionised water to a final volume of 25 μl. The reaction mixture was overlaid with sterile mineral oil and tubes placed on the heating plate of a Programmable Thermal Controller PTC-100, MJ Research Inc. Thermal cycling conditions were optimised to an initial denaturation at 94°C for 3 min, followed by 30 cycles of 30 s at 94°C (denaturation), 30 s at 53°C (annealing) and one minute at 72°C (extension). A final 5 min at 72°C ensured full extension of all amplified products. PCR products were resolved in a 3% Metaphor (FMC® BioProducts) high-resolution agarose matrix, using submarine gel electrophoresis following the supplier's instructions. Results were visualised by UV trans-illumination, after staining the gel with 0.5 μg/ml ethidium bromide. Alleles were detected and sized relative to a 100 bp ladder and standards of known size using a digital image of gels and the program UVIMAP (version 99 Software for Windows 95. UVItec, St John's Innovation Centre, Cambridge, UK).
Population genetic analysis was carried out using the program POPGENE (Yeh, 1998). For AFLP data, Nei and Shannon's diversity indices were calculated together with the number of polymorphic loci. For SSR data, observed allele number (No), allele frequency (fq), Shannon's diversity index (I), expected (He) and observed (Ho) heterozygosity were calculated. A χ2 test of significance for deviation from expected heterozygosity was carried out together with calculations of FST, FIS (Nei, 1987) and inferred gene flow [Nm, as 0.25(1−FST)/FST)]. For the AFLP data, the extent and significance of hierarchical partitioning of genetic variation among populations was calculated from nested genetic distance variance components (based on 1−S, where S is the distance estimate of Tanimoto, Deichsel and Trampisch, 1985) by the program WINAMOVA 1.55 (analysis of molecular variation; Excoffier et al, 1992). Significance values were assigned to variance components based on 1000 random permutations of individuals assuming no genetic structure.
Genetic/spatial autocorrelation of fine-scale population structure was examined using Moran's index for SSR data (Streiff et al, 1999) and Tanimoto's distance for AFLP data. The analysis was performed using the software package Spatial Genetic Software (SGS B. Degen, INRA Bordeaux). For each neighbourhood distance class, the observed Moran's index or Tanimoto distance (D(obs)), and a positive and a negative measure of the 95% confidence intervals (D (−CI) and D (+CI)) were provided. These latter values were based on 1000 resamplings of genotypes among the existing tree coordinates. If D(obs) is greater than the D(+CI), then there is a significant autocorrelation at that distance class and therefore significant genetic structure. A plot of Moran's index or Tanimoto's distance against neighbourhood distance classes, called a correlogram, allows for easy visualisation of genetic structure within a population.
The frequency of alleles in progeny arrays was compared to the mature tree generation for each of the sampled populations (Pocosol and Caño Negro). A simple exclusion approach was used to calculate the level of outcrossing from the SSR data for all progeny arrays.
Results
Genetic diversity
A total of 215 AFLP fragments were amplified across 110 S. macrophylla individuals, of which 108 were polymorphic (50.2%). The proportion of polymorphic loci and diversity indices for each population are presented in Table 2. Based on Nei's and Shannon's diversity estimates, Pocosol was the most diverse, followed by San Emilio and then Caño Negro; however, differences were not significant at the 95% level.
SSR analysis revealed a total of 18 alleles at three loci in the 73 mature trees surveyed from two populations (Caño Negro and Pocosol). Overall, the total number of alleles detected using the SSR primers of White and Powell (1997) was low compared to that found within a population of S. humilis, the species from which they were originally isolated. Often, when microsatellites are transferred to related species, the level of diversity can drop (Jarne and Lagoda, 1996), and the slightly lower resolving power of Metaphor gels (used in this study) compared to polyacrylamide (used by White and Powell, 1997) may also be a factor. The frequency of alleles differed markedly between the two surveyed populations (Table 3). For Caño Negro, each locus exhibited one allele at high frequency, ie, from 72%, at locus 54, to 91%, at locus 49. At Pocosol, however, several alleles exhibited intermediate frequencies and it was rare to have a single allele dominating. For example, the most frequent allele at locus 49 occurred in 36% of individuals, and for locus 54 it occurred in 68% of individuals. For the 36 individuals from Caño Negro, only two, three and three alleles were detected at the loci 49, 54 and 58, respectively. This is low in comparison with the 37 mature trees from Pocosol, for which six, five and four alleles were detected at the same loci, respectively. There were, on average, 2.66 observed alleles (No) for Caño Negro, and five observed alleles (No) for Pocosol. The Shannon's diversity index (I) shows that Pocosol (1.06±0.32) has almost twice the diversity of Caño Negro (0.55±0.24) and was significant at the 95% level (Table 3).
Population differentiation
Differentiation between populations, as measured by AFLP analysis, was relatively high (PhiST=0.383) with 38.3% of the variation being partitioned among populations (Table 4). All pairwise PhiST distances between populations were significant (P<0.001) and reflected geographic distance, with Caño Negro and San Emilio being more similar to each other (0.207) than either was to Pocosol (0.462 and 0.433, respectively).
Analysis of SSR data also indicated that the genetic differentiation between Pocosol and Caño Negro was fairly high (FST=0.24) and was of a similar order of magnitude to that calculated from the AFLP data. An indirect estimate of gene flow between the two populations was accordingly low (Nm=0.791). In addition, the mean observed heterozygosity at Caño Negro was significantly lower (Ho=14.8%) than expected (He=30.9% P<0.05; Table 5). No such relationship was observed at Pocosol where Ho (52.2%) was not significantly different from the expected value (He=58.4%; Table 5). A calculation of Wright's inbreeding coefficient for both populations indicated a heterozygosity deficit (FIS=0.24).
Fine-scale genetic structure
To examine the relation between genetic distance and tree proximity, a spatial autocorrelation analysis was performed for both the AFLP and SSR data from all sampled populations. Significant positive autocorrelation (ie, individuals within a distance class were more genetically similar than expected) was detected at the smallest distance classes within all populations. For Caño Negro, positive autocorrelation was detected at 0–50 m for AFLP and at 100 m for SSR; for Pocosol, at 0–100 m for AFLP and 0–50 m for SSR; and for San Emilio, at 0–100 m for AFLP. For all populations, significant negative autocorrelation (ie, individuals within the distance classes were less similar than expected) was evident within larger distance classes. For Caño Negro, negative autocorrelation was detected at 200 m for SSR (results were insignificant for AFLP); for San Emilio, at 250–300 m for AFLP; and for Pocosol at 300 m for SSR (results were insignificant for AFLP analysis although a similar trend was observed). Such a pattern indicates significant clumping of genetically similar individuals at all sites.
Outcrossing rate and gene flow
The overall power to discriminate potential fathers within progeny was significantly higher in Pocosol (70%) than in Caño Negro (39%), and is mainly because of diversity differences between the two populations. The minimum outcrossing rate, as calculated by simple exclusion, was 28.8% for Caño Negro and 50% for Pocosol. However, because of the low exclusion probabilities, this figure probably represents a large underestimate of the level of outcrossing in both populations, and thus should be considered a minimum level. Within progeny arrays and over the three SSR loci tested, mother trees in Pocosol sampled on average 0.75 nonself alleles (out of a total of six allelic loci available), whereas Caño Negro mother trees only sampled 0.33 nonself alleles. For both populations, none of the sampled progeny exhibited alleles that had not been previously identified within the sample of mature trees (n=37 and 36, respectively) surrounding mother trees (see Figure 1).
Discussion
Selective logging is expected to have a detrimental impact on the genetic resources of exploited timber tree species (Rodan et al, 1992; Newton et al, 1999). However, the response of different species to such external pressures is also expected to vary according to their biology and the intensity of impact. Here, we examined the fine-scale genetic structure and gene flow within disturbed populations of mahogany, S. macrophylla, a species for which a history of logging has been shown to be correlated with a reduction in population genetic diversity (Gillies et al, 1999).
Population differentiation and diversity
AFLP analysis indicated that the majority of genetic diversity is partitioned within (61.7%) rather than between S. macrophylla populations (PhiST=0.383). All populations were significantly differentiated from one another but Caño Negro and San Emilio were more similar to one another, than either was to Pocosol, reflecting their geographic location (Figure 1). Similarly, SSR analysis found that population subdivision between Caño Negro and Pocosol was significant (FST=0.24), and was reflected in the low level of inferred gene flow (Nm=0.79).
The overall estimates of population differentiation are similar to other outcrossing tree species (Hamrick et al, 1992), and to that of a previous RAPD study across the S. macrophylla range in Central America (Gillies et al, 1999), which indicated that 80% of variation is partitioned within populations. Compared to this estimate, differentiation between Pocosol and the other populations is high. Pocosol was also much more diverse than either of the other two populations. The central mountain range of Costa Rica is expected to present a partial barrier to gene flow and therefore increase differentiation over time. However, there are also significant environmental differences between the regions where Pocosol (low rainfall) and Caño Negro/San Emilio (high rainfall) populations are located. Similar patterns of differentiation and diversity have been noted for other species that inhabit both dry and moist habitats. Boshier and Lamb (1997), in their study of Cordia alliodora, found that populations occupying dry habitats were more diverse than, and genetically differentiated from, those in wet habitats. A similar pattern was also observed for Cedrela odorata, where dry and wet forms appear to be maintaining themselves as distinct and genetically differentiated ecotypes (Cavers and Lowe, in press). Both studies discuss the potential influence of glacial climate oscillations on the maintenance of diversity within dry-adapted ecotype populations. Within the genus Swietenia, the dry-adapted species, S. humilis, found primarily along the Pacific slopes of Central America, is differentiated from the more widespread S. macrophylla. The Pocosol population was identified on morphological grounds as S. macrophylla (C Navarro, unpublished). However, these results may indicate some level of ecotypic differentiation within S. macrophylla, or cryptic hybridisation with S. humilis in this area, and warrants further attention.
The link between population disturbance and a reduction in genetic diversity has been made for S. macrophylla (Gillies et al, 1999) and other species (eg Carapa guianensis, Hall et al, 1994). In addition to diversity differences, the mean observed (Ho) and expected heterozygosity levels (He) across all SSR loci were significantly higher for Pocosol (0.52 and 0.58, respectively) than for Caño Negro (0.15 and 0.31, respectively). The observed heterozygosity was also significantly lower than expected for Caño Negro, which was not the case for Pocosol. Such a result indicates a heterozygote deficit in Caño Negro that is not present at Pocosol. Two of the classical explanations for heterozygote deficit within populations are local population substructure and increased inbreeding. Boshier and Lamb (1997) found evidence of nonrandom mating in a predominantly outcrossing Costa Rican population of Cordia alliodora. Heterozygosity deficit was, however, explained by the local population structure. For the two S. macrophylla populations for which heterozygosity estimates were obtained, very similar patterns of fine-scale genetic structure existed (Figure 2), and thus population substructure is unlikely to be responsible. Hall et al (1994) found high levels of heterozygosity deficit in heavily logged populations of C. guianensis, and for which a causal link between breeding disruption and genetic structure was made. A study of fragmented Swietenia humilis populations (White et al, 1999) found that as fragment size decreased, diversity decreased (particularly the loss of low-frequency alleles) and heterozygote deficit (FIS) increased. It is possible therefore that recent heavy logging at Caño Negro, relative to the historical disturbance experienced at the Pocosol, could have caused a shift in the breeding system of mahogany at this site, and is reflected in the diversity and heterozygosity differences. To make predictive comparisons between the intensity of disturbance and the levels of diversity exhibited, a more detailed documentation on the intensity of extraction is required. To address this issue a further examination of the impact of logging and fragmentation within S. macrophylla will be necessary. In particular, it will be important to assess diversity, pre- and postimpact, within pairs of populations.
Fine-scale genetic structure
For all surveyed populations, similar levels of fine-scale structure were detected and indicate that clumps of genetically similar or related neighbourhoods are present. One explanation for such a uniform pattern of genetic structure across populations is seed-dispersal mechanism. Although an obligate outcrosser, S. macrophylla has heavy seeds that tend to fall close to the maternal tree. Within forests, seed dispersal for the species is limited and estimated to be between 32 and 80 m (Gullison et al, 1996). Such seed-dispersal distances would therefore be expected to establish half-sib neighbourhoods on a spatial scale of less than 100 m. Boshier and Lamb (1997) also found a spatial clustering of genetically similar individuals of C. alliodora within the first 50 m, a distance that coincides with its seed-dispersal range.
Outcrossing rate and gene flow
Minimum outcrossing rates, based on simple exclusion, were found to be 50% within Pocosol progeny arrays and only 28.8% at Caño Negro. However, the low exclusion probabilities most likely produced a major underestimation of outcrossing rate. Earlier findings of Mayhew and Newton (1998) indicate that the species is an obligate outcrosser with a form of self-incompatibility, and our data do not refute this assertion for either of the studied populations.
The overall level of pollen-mediated gene flow appears to be very limited, as all alleles present in seed families were found in trees that grew in the sample plot surrounding the mother trees (within approximately 200 m). Both sample plots are surrounded by continuous forest, where further sampling would certainly have uncovered additional allelic variation. Gene flow over a wide geographic area would therefore have been evident from the presence of nonproximate alleles within progeny arrays. Small insects, bees, moths and thrips are the main pollinators of S. macrophylla. Within a continuous forest canopy, these insects are expected to fly only short distances and thus the pollen-mediated gene flow estimates fit with the behavioural expectations for the predominant forest pollinators of S. macrophylla.
Another study of gene flow within populations of the closely related species, S. humilis (White, 1998), also indicated that within a large forest block most pollination events involve neighbouring trees (64%). However, as fragment size decreases, and separation between fragments increases, the number of long-distance pollination events increase (eg only 31.6% of pollinations involve neighbours within a block size of eight trees). White (1998) speculates that the windy conditions in open grassland increase gene flow between isolated fragments by blowing otherwise territorial, but weak flying, pollinators between forest blocks. Such nonintuitive consequences of fragmentation on gene diversity dynamics have also been noted for other species (eg Dinizia excelsa, Dick, 2001). However for Swietenia species, while long-distance gene flow is possible in a fragmented forest landscape, it was not found to be the case within continuous or disturbed forest.
Genetic resource management
Suitable genetic resource management strategies need to be developed in the context of the biology (eg regeneration ecology) and commercial pressures on a species. Mahogany has poor seed germination and seedling establishment within natural and disturbed forests (Mayhew and Newton, 1994), that is thought to be because of lack of light and overtopping by specialist gap colonists. The best conditions for the establishment of mahogany seedlings are in large deforested blocks, for instance following hurricanes or forest fires (Snook, 1998). Under such open conditions, mahogany seed can be dispersed long distances from trees in surrounding forest, and seedlings establish as large clusters of mahogany trees. Indeed, it has been argued that mahogany has evolved to be adapted to regenerate and propagate under such habitat dynamics within the hurricane-ravaged forests of Central America (Janzen, 1983; Snook, 1998). The clustering of genetically similar individuals observed in the surveyed Costa Rican forests could have beeen established following deforestation of large patches of forest (either by natural forest fires or following shifting cultivation). In addition, gene flow via pollen and seed appears to increase between fragmented forest patches relative to that within pristine or disturbed forest. In a similar way that selective logging inhibits the regeneration of mahogany seedlings, an intact or disturbed forest canopy also inhibits long-distance gene flow, and therefore also appears to restrict the recovery of genetic diversity in mahogany populations that have been exploited. Thus, it appears that a timber extraction strategy that harvests the entire blocks of forest (marketing both high and low value timbers and nontimber forest products) in rotation is best suited to sustain genetic diversity and promote regeneration of mahogany, and is being actively promoted as an alternative to selective logging strategies (Snook, 1998).
References
Barros PL, Queiroz WT, Silva JNM, Oliveira JNMF de A, Filho PPC, Terezo EF de M, Farias MM, Barros AV (1992). Natural and Artificial Reserves of Swietenia Macrophylla King in the Brazilian Amazon – a Perspective for Conservation. Faculdade de Ciencias Agropecuria: Belem, Para, Brazil.
Bawa KS, Perry DR, Beach JH (1985). Reproductive biology of tropical lowland rainforest trees. Sexual systems and incompatibility mechanisms. Am J Bot 72: 331–345.
Boshier DH, Lamb AT (1997). Mating system, population structure and gene flow. Cordia alliodora — genetics and tree improvement. Trop For Papers, 36: 29–36.
Cavers S, Lowe AJ (in press). Regional scale genetic structure within three Central American tree species; the influence of geography, biology and geological history. In: Degen B, Kremer A (eds) Modelling and Experimental Research on Genetic Processes in Tropical and Temperate Forests. INRA, EMBRAPA: Kourou, French Guyane.
Deichsel G, Trampisch HJ (1985). Clusteranalyse und Diskriminanzanalyse. Gustav Fischer Verlag: Stuttgart. pp 24.
Dick C (2001). Genetic rescue of remnant tropical trees by an alien pollinator. Proc R Soc Lond B 268: 2391–2396.
Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479–491.
Gillies ACM, Navarro C, Lowe AJ, Newton AC, Hernández M, Wilson J, Cornelius JP (1999). Genetic diversity in meso-American populations of mahogany (Swietenia macrophylla), assessed using RAPDs. Heredity 83: 722–732.
Goldstein DB, Schlötterer C (1999). Microsatellites. Evolution and Applications. Oxford University Press: Oxford.
Gullison RE, Paufil SN, Strouse JJ, Hubbell SP (1996). Ecology and management of mahogany (Swietenia macrophylla King) in the Chimanes Forest, Beni, Bolivia. Bot J Linn Soc 122: 9–34.
Hall P, Orrell LC, Bawa KS (1994). Genetic diversity and mating system in a tropical tree, Carapa guianensis (Meliaceae). Am J Bot 81: 1104–1111.
Hamrick JL, Godt MJW, Sherman-Broyles S (1992). Factors influencing levels of genetic diversity in woody plants species. New For 6: 95–124.
Janzen DH (1983). Costa Rican Natural History. University of Chicago Press: Chicago.
Jarne P, Lagoda PJL (1996). Microsatellites, from molecules to populations and back. Trends Ecol Evol 11: 424–429.
Mayhew JE, Newton AC (1998). The Silviculture of Mahogany. CABI Publishing Oxford: New York.
Myers N (1989). Deforestation Rates in Tropical Forests and their Climatic Implications. Friends of the Earth: London, England.
Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York. pp 159–164.
Newton AC, Cornelius JP, Baker P, Gillies ACM, Hernàndez M, Ramnarine R, Mesén JF, Watt AD (1996). Mahogany as a genetic resource. Bot J Linn Soc 122: 61–73.
Newton AC, Alnutt TR, Gillies ACM, Lowe AJ, Ennos RA (1999). Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14: 140–145.
Newton CR, Graham A (1994). PCR — Polymerase Chain Reaction. The Alden Press Ltd., Oxford, UK.
Pennington TD (1981). A Monograph of the Neotropical Meliaceae. Flora Neotropica. The New York Botanical Garden: New York. pp 396–399.
Rodan BD, Newton AC, Verissimo A (1992). Mahogany conservation: status policy and initiatives. Environ Cons 19: 331–338.
Snook LK (1998). Sustaining harvests of mahogany (Swietenia macrophylla King) from Mexico's Yucatán forests: past, present and future. In: Primack R, Galletti H, Ponciano I (eds) Timber, Tourists and Temples. Island Press: Washington, DC. pp 61–80.
Streiff R, Ducousso A, Lexer C, Steinkellner H, Glossl J, Kremer A (1999). Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Quercus petraea (Matt.) Liebl. Mol Ecol 8: 831–841.
Styles BT, Khosla PK (1976). Cytology and reproductive biology of Meliaceae. In: Burley J, Styles BT (eds) Tropical Trees: Variation, Breeding and Conservation. Linnean Society: London. pp 61–67.
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995). AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 23: 4407–4414.
Weising K, Nybom H, Wolff K, Meyer W (1995). DNA Fingerprinting in Plants and Fungi. CRC Press: London.
White G (1998). A Study of the Population Genetics of Swietenia humilis Zucc. In fragmented forest. PhD thesis, University of Dundee.
White G, Powell W (1997). Isolation and characterisation of microsatellite loci in Swietenia humilis (Meliaceae): an endangered tropical hardwood species. Mol Ecol 6: 851–860.
White G, Boshier DH, Powell W (1999). Genetic variation within a fragmented population of Swietenia humilis Zucc. Mol Ecol 8: 1899–1909.
Whitmore JL (1983). Swietenia macrophylla and S humilis (Caoba, mahogany). In: Janzen DH (ed) Costa Rica Natural History. University of Chicago Press: Chicago, London. pp 331–333.
Yeh F (1998). Population genetic analysis POPGENE Vers 1.31 (32-bit). Molecular Biology and Biotechnology Centre, University of Alberta, Canada and Centre for International Forestry Reseach, Indonesia.
Acknowledgements
We thank Marvin Hernandez and Lionel Coto for making field collections; Amanda Gillies and Jonathan Cornelius for providing constructive criticism to earlier drafts of the manuscript; and Antoine Kremer for helpful discussion. The financial support of NERC and the EU is gratefully acknowledged and this work was undertaken as part of an INCO-DC funded project ‘Assessment of levels and dynamics of intra-specific genetic diversity of tropical trees for conservation and sustainable management’, contract number ERBIC18CT970149.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lowe, A., Jourde, B., Breyne, P. et al. Fine-scale genetic structure and gene flow within Costa Rican populations of mahogany (Swietenia macrophylla). Heredity 90, 268–275 (2003). https://doi.org/10.1038/sj.hdy.6800247
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800247
Keywords
This article is cited by
-
Screening and identification of a Penicillium brevicompactum strain isolated from the fruiting body of Inonotus obliquus and the fermentation production of mycophenolic acid
Annals of Microbiology (2019)
-
Genetic diversity and population structure of Melia azedarach in North-Western Plains of India
Trees (2016)
-
Influence of putative forest refugia and biogeographic barriers on the level and distribution of genetic variation in an African savannah tree, Khaya senegalensis (Desr.) A. Juss
Tree Genetics & Genomes (2015)
-
Genetic analysis of the dry forest timber tree Sideroxylon capiri in Costa Rica using AFLP
Plant Systematics and Evolution (2015)
-
Molecular Markers and Conservation of Plant Species in the Latin-America: The Case of Phaedranassa viridiflora (Amaryllidaceae)
The Botanical Review (2013)