Abstract
Background:
The discovery of ERG/ETV 1 gene rearrangements and PTEN gene loss warrants investigation in a mechanism-based prognostic classification of prostate cancer (PCa). The study objective was to evaluate the potential clinical significance and natural history of different disease categories by combining ERG/ETV 1 gene rearrangements and PTEN gene loss status.
Methods:
We utilised fluorescence in situ hybridisation (FISH) assays to detect PTEN gene loss and ERG/ETV 1 gene rearrangements in 308 conservatively managed PCa patients with survival outcome data.
Results:
ERG/ETV1 gene rearrangements alone and PTEN gene loss alone both failed to show a link to survival in multivariate analyses. However, there was a strong interaction between ERG/ETV1 gene rearrangements and PTEN gene loss (P<0.001). The largest subgroup of patients (54%), lacking both PTEN gene loss and ERG/ETV1 gene rearrangements comprised a ‘good prognosis’ population exhibiting favourable cancer-specific survival (85.5% alive at 11 years). The presence of PTEN gene loss in the absence of ERG/ETV1 gene rearrangements identified a patient population (6%) with poorer cancer-specific survival that was highly significant (HR=4.87, P<0.001 in multivariate analysis, 13.7% survival at 11 years) when compared with the ‘good prognosis’ group. ERG/ETV1 gene rearrangements and PTEN gene loss status should now prospectively be incorporated into a predictive model to establish whether predictive performance is improved.
Conclusions:
Our data suggest that FISH studies of PTEN gene loss and ERG/ETV1 gene rearrangements could be pursued for patient stratification, selection and hypothesis-generating subgroup analyses in future PCa clinical trials and potentially in patient management.
Similar content being viewed by others
Main
Prostate cancer (PCa) is the most commonly diagnosed male cancer and the second commonest cause of male cancer related mortality in the Western world (Ferlay et al, 2007). The clinical behaviour and molecular pathology of PCa is highly variable. There is an urgent need to dissect this inter-patient heterogeneity with robust molecular biomarkers to accelerate the successful conduct of clinical trials for this disease, optimise patient treatment and minimise late drug development attrition (Betensky et al, 2002; Attard et al, 2008a). Critically, identifying patient subgroups that require less treatment from those that should be targeted with more aggressive therapy is a key goal.
PTEN loss and ETS gene rearrangements are proposed to be critically important and common molecular events in prostate carcinogenesis (Trotman et al, 2003; Tomlins et al, 2005, 2008a; Carver et al, 2009; King et al, 2009). In particular, recent publications have addressed the relationship between the two events in mouse models demonstrating cooperation (Carver et al, 2009; King et al, 2009). Deletion of all or part of the tumour suppressor gene PTEN is a frequent event. Other classes of alterations including mutations and post-translational modifications occur less frequently (Whang et al, 1998; Verhagen et al, 2006; Yoshimoto et al, 2007, 2008). Previous studies have examined the prognostic significance of PTEN deletions by fluorescence in situ hybridisation (FISH) with small patient cohorts and biochemical recurrence as the outcome (Yoshimoto et al, 2007, 2008). Similarly the clinical implications of the ETS gene rearrangements (Tomlins et al, 2005, 2007) are not yet fully understood (Clark and Cooper, 2009), with their presence reported as associated with both poor (Demichelis et al, 2007; Nam et al, 2007; Attard et al, 2008b) and good prognosis (Petrovics et al, 2005; Gopalan et al, 2009; Hermans et al, 2009). We reported that deletion of the intermediate region between TMPRSS2 and ERG combined with duplication of the TMPRSS2–ERG fusion sequences is predictive of poor cancer-specific survival (Attard et al, 2008b), an observation supported by other studies (FitzGerald et al, 2008; Yoshimoto et al, 2008; Gopalan et al, 2009). The effect of PTEN loss on PCa survival and its relationship to the ETS gene rearrangements is largely unknown.
To evaluate the natural history of the different disease variants identified by PTEN, ERG and ETV1 FISH we evaluated a retrospective cohort of conservatively managed men (Cuzick et al, 2006). Immunohistochemistry (IHC) was not carried out because of concerns regarding potential interobserver interpretation variability (Kirkegaard et al, 2006); lack of uniformity regarding tissue processing as samples were taken from different hospitals (Cuzick et al, 2006); and previous studies reporting conflicting results with IHC (Bedolla et al, 2007; McCall et al, 2008; Sircar et al, 2009). Rearrangements involving ETV4 and ETV5 have also been reported, but these are rare and therefore unlikely to be used widely in a clinically applicable test (Kumar-Sinha et al, 2008).
Patients and methods
Patient cohort
Tissue microarrays (TMAs) were constructed from unselected transurethral resection of prostate (TURP) specimens from patients who received no initial treatment in a cohort of conservatively managed men with PCa (Cuzick et al, 2006). Ethical approval for the collection of the cohort was obtained from the Ethics Review Committees at every collaborating hospital.
TMAs and FISH studies
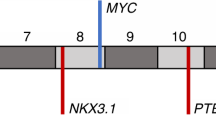
Tissue microarrays were constructed as previously described (Attard et al, 2008b). Areas of ‘cancer’ and ‘normal’ were identified on the basis of histopathological examination of haematoxylin and eosin and p63/AMACR-stained sections that flanked the TMA slice used for FISH studies (Figure 1). To assess the frequency of PTEN gene loss in TMA cancer cores, FISH procedures were carried out as described previously for rearrangement at the ERG and ETV1 loci (Attard et al, 2008b, 2008c). The same (or an immediately adjacent) slice was stripped and rehybridised for PTEN. We selected PTEN BACs that map to the minimum region of PTEN deletion in PCa as previously described in xenografts and cell lines (Hermans et al, 2004). We used two, overlapping, DIG-labelled BAC probes to the 5′ end of the PTEN locus, RP11-765C10 and RP11-959L24, and a commercially available CY-3 labelled DNA chromosome 10 centromere probe (chromosome 10 (p11.1∼q11.1 Abbott Molecular, Des Plaines, IL, USA) (Figure 2A). Representative images of nuclei with normal, heterozygous and homozygous PTEN patterns are shown in Figures 1 and 2B.
Haematoxylin and eosin (H & E), fluorescence in-situ hybridisation (FISH) and P63/alpha-methylacyl-CoA racemase (AMACR) on adjacent slides (A) a prostate cancer gland with H & E staining; (B) ETV1 FISH in the same gland on an adjacent slide. Cartoon and magnified images of two nuclei are also shown. The upper nucleus has four paired (ploidy) ETV1 probes and the lower nucleus has two paired ETV1 probes indicating wild-type ETV1; FISH for ERG also showed paired probes indicating wild-type ERG (image not shown); (C) PTEN FISH in the same gland on an adjacent slide. Cartoon and magnified images of one nucleus is shown. The nucleus has four (ploidy) chromosome, 10 centromeric probes (in red) and two PTEN probes (in green) indicating heterozygous loss of PTEN; and (D) the same prostate cancer gland on an adjacent slide, which has absent P63 staining and AMACR positivity.
Fluorescence in-situ hybridisation (FISH) detection of PTEN loss. (A) Position of two ‘PTEN’ BAC probes shown in green. The PTEN gene and flanking gene C10orf59 are shown in dark blue. Arrows indicate the direction of transcription. (B) PTEN loss patterns. Green signals are probes that detect PTEN and red signals are probes that detect the chromosome 10 centromere. Nuclei with normal PTEN complement are visualised in interphase as two green and two red signals (left). Heterozygous PTEN loss results in the loss of one green signal (centre) and homozygous PTEN loss, the loss of two green signals (right).
Establishing PTEN cut-offs
Evaluation of the FISH results in each core containing cancer was independently carried out by two operators (A-R and G-A) who were unaware of clinical data. Signals were scored in at least 200 non-overlapping nuclei. We assumed that part of some nuclei can be lost during slicing and therefore not all nuclei in a section would have their full probe complement. Using accepted methodology described previously we estimated the degree of technical artefact by studying nuclei in TMA cores with non-malignant prostatic epithelium (Ventura et al, 2006). As cancer nuclei may differ in size from non-malignant nuclei, we also counted patterns of PTEN loss in cancer nuclei for which an overall ‘normal PTEN complement’ FISH score had been given. These data are provided in Supplementary Figure 1 and Supplementary Table 1.
Statistical analysis
Associations between PTEN status and categorical data were examined using the χ2 test for trend. Associations between PTEN status and numerical variables were assessed using analysis of variance. The primary endpoints for this study were time to death from PCa and time to death from any cause. Univariate and multivariate analyses were carried out by proportional hazard (Cox) regression analysis (Cox and Oakes, 1984). The following variables, determined as described previously (Cuzick et al, 2006), were included in the multivariate analyses: centrally reviewed Gleason scores determined by modern grading criteria, baseline PSA (PSA within 6 months of diagnosis) and age at diagnosis. All P-values were two-sided and all statistical analyses were carried out using Stata 10.0 (StataCorp, LP, College Station, TX, USA).
Results
General
PTEN gene status was established from 678 cancer cores from 322 patients. Tissue cores can hybridise with variable efficiency for different FISH probes. Moreover, cores can sometimes be lost in the rehybridisation process. Therefore, ERG and ETV1 gene status was available from 308 of the 322 patients with a PTEN score (662 TMA cancer cores). Demographics and characteristics for tumours stratified for PTEN status alone are shown in Table 1 and according to PTEN and ERG/ETV1 gene status shown in Table 2. The median follow-up was 100 months (3–197 months). A competing risk analysis demonstrated that after 11 years of follow-up, 59% of men had died: 25% from PCa and 34% from other causes; only 22% were alive without progression. Results are described for both cause-specific prostate survival and overall survival.
PTEN gene loss alone is not a significant predictor of clinical outcome in multivariate analysis
There was no statistically significant difference in outcome between the tumours with heterozygous and those with homozygous PTEN loss (data not shown). Therefore, the analyses presented consider PTEN loss tumours as one group. Of the 322 patients with a PTEN score, 266 (83%) had a normal PTEN score and 56 (17%) had PTEN loss. There were significant associations between PTEN loss and Gleason score (P<0.001), clinical stage (P<0.001), baseline PSA (P<0.001) and cancer in biopsy (proportion of TURP chips with disease or linear proportion of needle biopsy containing disease) (P<0.001), but no association with age (Table 1). Univariate Cox analysis demonstrated that compared to cancers with PTEN, tumours with PTEN loss had significantly worse cause-specific and overall survival (HR=3.33, 95% CI=2.11–5.26, P<0.001, Figure 3A, and HR=1.72, 95% CI=1.24–2.38, P=0.001). However, in a multivariate model, PTEN loss did not retain significance as a prognostic factor for PCa survival (HR=1.19, 95% CI=0.73–1.96, P=0.49) or overall survival (HR=1.00, 95% CI=0.70–1.44, P=0.99). Furthermore, even when Gleason score only was added to the model, PTEN did not remain a significant independent prognostic factor for PCa survival (HR=1.61, 95% CI=0.99–2.62, P=0.05) or overall survival (HR=1.16, 95% CI=0.82–1.65, P=0.41).
PTEN gene loss and ERG/ETV 1 gene rearrangements
Univariate Cox analysis demonstrated that ERG/ETV1 gene rearrangement status alone was a significant prognostic factor for cause-specific and overall survival (HR=2.17, 95% CI=1.39–3.40, P=0.001 and HR=1.58, 95% CI=1.20–2.08, P=0.001). However, on multivariate Cox analysis ERG/ETV1 gene rearrangement status alone was not a significant independent prognostic factor for either cause-specific or overall survival (HR=0.93, 95% CI=0.58–1.50, P=0.78 and HR=1.04, 95% CI=0.77–1.41, P=0.80). We next considered PTEN and ERG/ETV1 gene status together (Table 2). The 308 patients were stratified into those that had an ERG or ETV1 gene rearrangement in their cancers (122 patients, 40%) and those that did not (186 patients, 60%). The patients were then further stratified as to whether they had PTEN loss or not. There was a significant association between PTEN score and ERG/ETV 1 status (P<0.001) with 66% of PTEN loss tumours also having an ERG/ETV1 gene rearrangement compared with 34% of normal PTEN tumours (Table 3). When the patients were stratified according to their ERG/ETV1 gene status there was a significant interaction with PTEN status (test for heterogeneity (Q (1df)=20.7, P<0.001 and Q (1df)=20.9, P<0.001 in multivariate analysis for cause-specific survival and overall survival, respectively). Cox analyses were therefore conducted using the group with normal ERG/ETV 1 and normal PTEN as the reference.
PTEN gene loss with no ERG/ETV 1 rearrangement identifies a poor prognosis group
Univariate analysis demonstrated that patients with normal ERG/ETV1 and PTEN loss had a significantly worse cause-specific and overall survival (HR=9.37, 95% CI=4.68–18.76, P<0.001, Figure 3B and HR=3.14, 95% CI=1.89–5.23, P<0.001 respectively). In multivariate analysis cases with normal ERG/ETV1 and PTEN loss had a significantly higher risk of dying from PCa (HR=4.87, 95% CI=2.28–10.41, P<0.001). The cancer-specific survival at 11 years was 13.7%. Similar results were found for overall survival (HR=2.40, 95% CI=1.40–4.11, P=0.001. Gleason grades of these tumours (Table 2) demonstrate that in 21% of the normal ERG/ETV1 and PTEN loss tumours, Gleason grade was <7 supporting a reclassification of this low Gleason grade patient subgroup.
ERG/ETV 1 gene rearranged tumours with and without PTEN loss form two intermediate prognostic groups
In univariate analysis men with rearranged ERG/ETV1 and normal PTEN exhibited significantly worse cause-specific and overall survival (HR=2.99, 95% CI=1.70–5.25, P<0.001, Figure 3B and HR=1.80, 95% CI=1.30–2.47, P<0.001 respectively) compared with men with normal ERG/ETV1 and PTEN status. Similar results were observed for cases with rearranged ERG/ETV1 and PTEN loss (HR=3.92, 95% CI=2.06–7.48, P<0.001, Figure 3B and HR=1.77, 95% CI=1.16–2.70, P=0.008, respectively). In multivariate analysis, men with rearranged ERG/ETV1 and normal PTEN had a marginally higher risk of dying from PCa (HR=1.82, 95% CI=1.01–3.26, P=0.04), but no effect was observed in cases with rearranged ERG/ETV1 and PTEN loss (HR=0.98, 95% CI=0.49–1.97, P=0.96). Similar results were found for overall survival (HR=1.37, 95% CI=0.98–1.92, P=0.06 and HR=0.84, 95% CI=0.52–1.35, P=0.48, respectively). The cancer-specific survival at 11 years for the rearranged ERG/ETV1 and normal PTEN group was 59.8% and for the rearranged ERG/ETV1 and PTEN loss group was 41.0% (Figure 3B).
No ERG/ETV 1 gene rearrangement and no PTEN gene loss identifies a good prognosis group
The largest group of patients (n=167, 54%) comprised those who had neither an ERG/ETV1 gene rearrangement nor PTEN loss. This group (mean age 69±5 years) had a greater cause specific survival (85.5% at 11 years) and overall survival when compared with the three other groups (Figure 3B). In this good prognosis group, 60 out of 167 (36%) had a Gleason grade ⩾7. Prostate cancer-specific deaths in this cohort were not confined to the higher Gleason grades (Table 2). Of these 20 men who died from PCa at 11 years, 5 (25%) had a Gleason score of <7; 7 (35%) had a Gleason score of 7; and the remaining 8 (40%) had a Gleason score of >7.
Discussion
We present the first large series in which PTEN and ERG/ETV1 gene status have been analysed together. We have identified patient subgroups with high and low risk of death from PCa based on PTEN and ERG/ETV1 status. There was no difference in outcome between tumours with heterozygous and those with homozygous PTEN loss and the PTEN loss tumours were therefore considered as one group. This may be because in heterozygous tumours by FISH, the other allele is lost by an alternative mechanism (Whang et al, 1998; Verhagen et al, 2006). The ‘good prognosis’ group (54%) lacked an ERG/ETV1 gene rearrangement and PTEN gene loss (85.5% PCa survival at 11 years). The PCa-specific deaths in this group did not only occur in the patients with the higher Gleason grades, but across the Gleason grades. These results highlight some inadequacy of Gleason grading in determining which patients require more intensive therapy for their PCa.
We also identified a patient group with a significant ‘poor prognosis’. Patients lacking an ERG/ETV1 gene rearrangement but with PTEN gene loss had the worst cause-specific survival of 13.7% at 11 years. A proportion of patients in this group had a Gleason score of ⩽7. These data are also potentially of clinical importance as they identify a patient group who could be targeted to receive more intensive neoadjuvant and adjuvant therapy when other clinicopathological parameters recommended a more conservative approach. This requires testing in prospectively designed studies. Agents that specifically target the PI3K–AKT–mTOR pathway are undergoing investigation in clinical trials (Yap et al, 2008) and future studies should specifically evaluate these agents in this subgroup.
One previous study (82 patients) reported that the absence of PTEN loss and no ERG gene rearrangement is associated with a longer time to biochemical recurrence and PTEN loss plus an ERG gene rearrangement was associated with the shortest time to biochemical recurrence (Yoshimoto et al, 2008). Our results are similar in that those tumours that had neither an ERG gene rearrangement nor PTEN loss were in a good prognostic group. However, we did not show that the combination of the two alterations was associated with poor outcome, and rather that it is those tumours with PTEN loss but no ERG/ETV1 gene rearrangement that do particularly badly. These differences may be a consequence of different study sizes or endpoints used.
ERG/ETV1 gene rearrangement positive and negative tumours have been reported to have distinct chromosomal aberrations, expression signatures and morphological features (Kumar-Sinha et al, 2008), thus suggesting that they represent different PCa classes. Our results support this hypothesis because PTEN loss was only a significant independent prognostic factor for overall survival, when analysed in the ERG/ETV1 gene non-rearranged tumours. These results also raise the question of whether there are other molecular abnormalities that are mutually exclusive of ERG/ETV1 gene rearrangements that may contribute to the worse outcome of patients with non-ERG/ETV1 gene rearranged/PTEN loss tumours. In this respect, the recent observation that a proportion of ETS-gene rearrangement negative cancers overexpress SPINK1 protein is of particular interest (Tomlins et al, 2008b). SPINK1 expression was linked to poorer outcome (Tomlins et al, 2008b) and SPINK1 has been shown, when overexpressed in colorectal and breast cancer cells, to function as an autocrine growth factor that can stimulate the PI3K pathway (Gouyer et al, 2008).
Conclusions
In conclusion, these data suggest that molecular characterisation of PTEN, ERG and ETV1 gene status might be used in future to determine the risk of PCa death. This has implications both for potentially deciding which patients should be conservatively or aggressively treated and also for stratification of patients in clinical trials. At present clinical trial patients are stratified by clinicopathological features alone. Our results suggest that an imbalance in numbers of patients with different PTEN and ERG/ETV1 gene status in different study arms could falsely influence trial outcome and needs to be accounted for.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Attard G, Ang JE, Olmos D, de Bono JS (2008a) Dissecting prostate carcinogenesis through ETS gene rearrangement studies: implications for anticancer drug development. J Clin Pathol 61: 891–896
Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter V, De Bono JS, Scardino P, Cuzick J, Cooper CS (2008b) Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene 27: 253–263
Attard G, Clark J, Ambroisine L, Mills IG, Fisher G, Flohr P, Reid A, Edwards S, Kovacs G, Berney D, Foster C, Massie CE, Fletcher A, De Bono JS, Scardino P, Cuzick J, Cooper CS (2008c) Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer 99: 314–320
Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, Ghosh PM (2007) Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res 13: 3860–3867
Betensky RA, Louis DN, Cairncross JG (2002) Influence of unrecognized molecular heterogeneity on randomized clinical trials. J Clin Oncol 20: 2495–2499
Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP (2009) Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 41: 619–624
Clark JP, Cooper CS (2009) ETS gene fusions in prostate cancer. Nat Rev Urol 6: 429–439
Cox D, Oakes D (1984) Analysis of Survival Data. Chapman and Hall: London, New York
Cuzick J, Fisher G, Kattan MW, Berney D, Oliver T, Foster CS, Moller H, Reuter V, Fearn P, Eastham J, Scardino P (2006) Long-term outcome among men with conservatively treated localised prostate cancer. Br J Cancer 95: 1186–1194
Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, Adami HO, Mucci LA, Kantoff PW, Andersson SO, Chinnaiyan AM, Johansson JE, Rubin MA (2007) TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene 26: 4596–4599
Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P (2007) Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol 18: 581–592
FitzGerald LM, Agalliu I, Johnson K, Miller MA, Kwon EM, Hurtado-Coll A, Fazli L, Rajput AB, Gleave ME, Cox ME, Ostrander EA, Stanford JL, Huntsman DG (2008) Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: results from a population-based study of prostate cancer. BMC Cancer 8: 230
Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, Eastham JA, Scardino PT, Scher HI, Tickoo SK, Reuter VE, Gerald WL (2009) TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res 69: 1400–1406
Gouyer V, Fontaine D, Dumont P, de Wever O, Fontayne-Devaud H, Leteurtre E, Truant S, Delacour D, Drobecq H, Kerckaert JP, de Launoit Y, Bracke M, Gespach C, Desseyn JL, Huet G (2008) Autocrine induction of invasion and metastasis by tumor-associated trypsin inhibitor in human colon cancer cells. Oncogene 27: 4024–4033
Hermans KG, Boormans JL, Gasi D, van Leenders GJ, Jenster G, Verhagen PC, Trapman J (2009) Overexpression of prostate-specific TMPRSS2(exon 0)-ERG fusion transcripts corresponds with favorable prognosis of prostate cancer. Clin Cancer Res 15: 6398–6403
Hermans KG, van Alewijk DC, Veltman JA, van Weerden W, van Kessel AG, Trapman J (2004) Loss of a small region around the PTEN locus is a major chromosome 10 alteration in prostate cancer xenografts and cell lines. Genes Chromosomes Cancer 39: 171–184
King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, Gerald WL, Sawyers CL (2009) Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet 41: 524–526
Kirkegaard T, Edwards J, Tovey S, McGlynn LM, Krishna SN, Mukherjee R, Tam L, Munro AF, Dunne B, Bartlett JM (2006) Observer variation in immunohistochemical analysis of protein expression, time for a change? Histopathology 48: 787–794
Kumar-Sinha C, Tomlins SA, Chinnaiyan AM (2008) Recurrent gene fusions in prostate cancer. Nat Rev Cancer 8: 497–511
McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J (2008) Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer 99: 1296–1301
Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, Stanimirovic A, Encioiu E, Neill M, Loblaw DA, Trachtenberg J, Narod SA, Seth A (2007) Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer 97: 1690–1695
Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Chen Y, Dobi A, Srikantan V, Sesterhenn IA, McLeod DG, Vahey M, Moul JW, Srivastava S (2005) Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 24: 3847–3852
Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen G, Darnel AD, Aprikian AG, Saad F, Bismar TA, Squire JA (2009) PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol 218 (4): 505–513
Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM (2007) Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448: 595–599
Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM (2008a) Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 10: 177–188
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648
Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, Demichelis F, Helgeson BE, Laxman B, Morris DS, Cao Q, Cao X, Andren O, Fall K, Johnson L, Wei JT, Shah RB, Al-Ahmadie H, Eastham JA, Eggener SE, Fine SW, Hotakainen K, Stenman UH, Tsodikov A, Gerald WL, Lilja H, Reuter VE, Kantoff PW, Scardino PT, Rubin MA, Bjartell AS, Chinnaiyan AM (2008b) The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell 13: 519–528
Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, Khoo AS, Roy-Burman P, Greenberg NM, Van Dyke T, Cordon-Cardo C, Pandolfi PP (2003) Pten dose dictates cancer progression in the prostate. PLoS Biol 1: E59
Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, Siebert R (2006) FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn 8: 141–151
Verhagen PC, van Duijn PW, Hermans KG, Looijenga LH, van Gurp RJ, Stoop H, van der Kwast TH, Trapman J (2006) The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol 208: 699–707
Whang YE, Wu X, Suzuki H, Reiter RE, Tran C, Vessella RL, Said JW, Isaacs WB, Sawyers CL (1998) Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci USA 95: 5246–5250
Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P (2008) Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol 8: 393–412
Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, Squire JA (2007) FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer 97 (5): 678–685
Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, Zielenska M, Soares FA, Squire JA (2008) Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol 21: 1451–1460
Acknowledgements
This work was funded by the National Cancer Research Institute, The Prostate Cancer Charity, The Grand Charity of Freemasons, The Rosetrees Trust, The Bob Champion Cancer Trust and The Royal Marsden Clinical Research Fund. Dr DM Berney is supported by The Orchid Appeal. Dr JS de Bono was supported by Cancer Research UK, the Medical Research Council, and Prostate Cancer Research Foundation. Dr G Attard is a Prostate Cancer Foundation Young Investigator and Drs Reid and Attard have received Cancer Research UK funding.
Originality and Research Support: I declare that the content of the manuscript is original and that it has not been published or accepted for publication, either in whole or in part, in any form. No part of the manuscript is currently under consideration for publication elsewhere.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Reid, A., Attard, G., Ambroisine, L. et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer 102, 678–684 (2010). https://doi.org/10.1038/sj.bjc.6605554
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605554
Keywords
This article is cited by
-
Capivasertib combines with docetaxel to enhance anti-tumour activity through inhibition of AKT-mediated survival mechanisms in prostate cancer
British Journal of Cancer (2024)
-
Taraxasterol suppresses the proliferation and tumor growth of androgen-independent prostate cancer cells through the FGFR2-PI3K/AKT signaling pathway
Scientific Reports (2023)
-
Characterization of exposure–response relationships of ipatasertib in patients with metastatic castration-resistant prostate cancer in the IPATential150 study
Cancer Chemotherapy and Pharmacology (2022)
-
Alterations of PTEN and SMAD4 methylation in diagnosis of breast cancer: implications of methyl II PCR assay
Journal of Genetic Engineering and Biotechnology (2021)
-
Loss of EGR3 is an independent risk factor for metastatic progression in prostate cancer
Oncogene (2020)