Abstract
Background:
We recently identified and validated UBE2C RNA as a prognostic marker in 252 node-positive (N+) breast cancers by means of a microarray study. The aim of this study was to validate UBE2C protein as a prognostic marker in N+ breast cancer by immunohistochemistry (IHC).
Methods:
To this end, 92 paraffin-embedded blocks were used. The impact of UBE2C IHC value on metastasis-free survival (MFS) and overall survival (OS) was evaluated and compared with Ki-67 and Nottingham prognostic index (NPI) performances.
Results:
In accordance with genomic data, UBE2C IHC had a significant impact both on MFS and OS (hazard ratio=6.79 – P=0.002; hazard ratio=7.14 – P=0.009, respectively). Akaike information criterion proved that the prognostic power of UBE2C IHC was stronger than that of Ki-67 (and close to that of NPI). Furthermore, multivariate analyses with NPI showed that, contrary to Ki-67 IHC, UBE2C IHC remained an independent factor, both for MFS (adjusted P=0.02) and OS (adjusted P=0.04).
Conclusion:
We confirmed that UBE2C protein measured by IHC could be used as a prognostic marker in N+ breast cancer. The potential predictive interest of UBE2C as a marker of proteasome activity needs further investigations.
Similar content being viewed by others
Main
The ubiquitin–proteasome system (UPS) is involved in many, if not all, cellular events through the regulation of protein homoeostasis and fate (Hershko et al, 2000). It consists of a key protein, ubiquitin; several enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (E2), ubiquitin-ligating enzymes (E3), deubiquitinating enzymes and a highly sophisticated protease complex, the 26S proteasome. The latter is made up of two subcomplexes: a catalytic core particle (20S proteasome: PSMA and PSMB subunits) and one or two terminal 19S particle(s) (PSMC and PSMD subunits), which serve as proteasome activators (Tanaka, 2009). With the multitude of substrates targeted and the numerous processes involved, it is not surprising that aberrations in this pathway have been implicated in the pathogenesis of many diseases, such as cancer and, more specifically, breast cancer (Lipkowitz, 2002; Ohta and Fukuda, 2004; Mani and Gelman, 2005).
Proteomic and genomic studies conducted by our team pointed out different elements of the UPS as breast cancer prognostic markers: ubiquitin as a prognostic protein marker in node-negative (N-) patients, and UBE2C, PSMA5, PSMB3, PSMB7, and PSMD3 as prognostic nucleic acid markers in node-positive (N+) patients, respectively (Ricolleau et al, 2006; Campone et al, 2008; Jézéquel et al, 2008). Our results were consistent with those of other studies (Chen and Madura, 2005; Deng et al, 2007). Of the 38 genes included in our 38-gene expression signature, UBE2C was the most highly ranked gene. Furthermore, evaluation of the prognostic power of this gene on external microarray data (more than one thousand) confirmed the robustness of this marker at the RNA level (Jézéquel et al, 2008). This finding was consistent with reports that underlined a strong link between UBE2C overexpression and the degree of tumour differentiation in many cancers (breast, lung, ovary, bladder, and glioblastomas) (Okamoto et al, 2003; Wagner et al, 2004). Furthermore, in breast cancer, an increased expression of UBE2C was associated with high tumour grade and cancer progression (Ma et al, 2003). Finally, UBE2C belongs to proliferative genes, which are known to constitute the majority of genes included in prognostic gene-expression signatures (Desmedt et al, 2008; Wirapati et al, 2008). The robustness of this marker may render it suitable for routine use, but the practicability of its measurement might be improved by using immunohistochemistry (IHC) staining. This technique might be preferred over nucleic acid measurement because of three advantages. First, IHC is considered as a practical approach in routine testing because of its relative inexpensiveness and straightforwardness, and is well established in standard clinical pathology laboratories. Second, IHC is applicable to paraffin-embedded tissues; there is no need for fresh, frozen, nonfixed tissue as a preferential requirement for nucleic acid quantification. Third, a more likely relevant validation consists of studying the expression level of a potential biomarker at a protein level, such as with IHC. Studies have shown that there is often discordance between levels of nucleic acids and proteins, implying that the study of both measures is important.
So the aim of this study was (1) to determine the prognostic power of UBE2C by means of IHC staining in 92 N+ breast cancer patients; (2) to compare the power of this proliferation marker with that of the reference marker in breast cancer, Ki-67, and (3) to evaluate if UBE2C added predictive accuracy to that provided by the combined prognostic index: Nottingham Prognostic Index (NPI) (Stuart-Harris et al, 2008).
Materials and methods
Patients
The study included paraffin-embedded blocks from 92 consecutive women with primary N+ breast tumours, who were diagnosed and treated primarily between April 1997 and July 2001 at the René Gauducheau Cancer Centre. Forty-three patients were also part of a previous genomic study that led to the identification of a 38-gene expression node-positive prognostic signature (Jézéquel et al, 2008). The median age at diagnosis was 51 years (range, 27–74 years). Informed consent was obtained from patients to use their surgical specimens and clinicopathological data for research purposes, as required by the French Committee for the Protection of Human Subjects. These patients showed no evidence of distant metastasis at the time of diagnosis. None had received chemotherapy, endocrine therapy or radiation therapy before surgery. Treatment decisions were based solely on consensus recommendations at the time of diagnosis. Patients were followed up for metastasis-free survival (MFS) (delimited by the first clinically recognised evidence of distant recurrence). All patients received FEC adjuvant chemotherapy and post-operative radiation therapy. Seventy-four received hormonotherapy (tamoxifen). Patients were followed up every 4 months during 2 years, then every 6 months during 3 years, and annually thereafter. Clinical examination, mammography and chest radiography were performed twice a year, and bone scintigraphy and liver ultrasonography annually.
IHC
Sections (3 μm) from formalin-fixed, paraffin-embedded tumours were cut and mounted on Superfrost Plus slides (VWR International, Leicestershire, UK). Following deparaffinisation in xylene, slides were rehydrated through a graded series of alcohol and placed in running water. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide and methanol. Samples were steamed before incubation for antigen retrieval with 10 mM citrate buffer (pH 6.0) for UBE2C and EDTA pH 7.2 for Ki-67. Slides were incubated for UBE2C (Boston Biochem, Cambridge, UK, dilution: 1/500) and Ki-67 (Dako, Glostrup, Denmark, clone MIB1, dilution: 1/100) using a biotin-streptavidin-peroxidase detection system (Kit ChemMate, Dako). 3,3′-diaminobenzidine tetrahydrochloride (DAB) was used for the visualisation of the antibody/enzyme complex. Slides were counterstained with haematoxylin. Negative controls were included in each case by omitting the primary antibody. Ki-67 and UBE2C scores were defined as the percentage of immunostained cells divided by the total number of cells in the evaluated area. All counts were performed at a magnification of × 40 using a standard light microscope. The percentage of UBE2C and Ki-67 stained cells was evaluated individually and independently by two pathologists (DL, FL) in a double-blind manner. For each case, 250 cells were counted.
Statistical analysis
Descriptive statistics
Categorical data were presented as frequencies and continuous variables were expressed as the median (range). It was also decided not to use the best, but most likely overfitted, cut-off for Ki-67 but the one calculated from 2685 patients by Viale et al (2008) 11% . The same one was used for UBE2C. Relationships between UBE2C IHC groups (<11% vs ⩾11%) and other parameters were determined by using the non-parametric Mann–Whitney test if continuous and Fisher's exact test if discrete.
Inter-observer reproducibility
Agreement between pathologists (DL and FL) for UBE2C and Ki-67 was verified in two ways: if continuous, by means of Spearman's correlation; if discretised (<11% vs ⩾11%), by means of Kappa test.
Correlation study between genomic and IHC data
Microarray and IHC data were available for 40 patients. Microarray characteristics and data have been deposited in the NIH Gene Expression Omnibus (Series accession number: GSE11264) according to minimum information about a microarray experiment (MIAME) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11264) (Jézéquel et al, 2008). UBE2C RNA expression was measured by means of the two different cDNA probes named: UMGC_02270 and UMGC_06429. Correlation between UBE2C RNA expression and UBE2C IHC was determined by means of Spearman's test. Moreover, to be able to extrapolate results concerning UBE2C IHC impact on MFS, the previously shown prognostic value of UBE2C genomic value on n=252 patients was verified on this subsample (n=40) by univariate Cox regression. UBE2C genomic data are detailed in Supplementary Table 1 (Jézéquel et al, 2008).
Survival analysis
Categorical time from surgery to metastasis relapse (MR) (primary end point) was retained for the study. Overall survival (OS), defined as time from surgery to death from any cause, was used to reinforce MFS analysis. MFS curves were plotted according to Kaplan–Meier method and compared by means of the log-rank test. Cox's proportional hazard regression analyses were performed on UBE2C and Ki-67 expression (discretised and continuous) and on all other known prognostic parameters to assess their independent association with MFS and OS. Proportional hazard assumption was verified for the final models by means of Schoenfeld residuals study. On account of the moderate number of MR and deaths during follow-up (n=25/92 and 18/92, respectively), permutation tests were performed both at univariate and multivariate steps to optimise the robustness of the results.
Comparison and independence
Prognostic power for MFS and OS was compared for UBE2C IHC and Ki-67 IHC both discretised at retained cut-off (<11% vs ⩾11%) and for the NPI (classical prognostic reference) by means of Akaike information criterion (AIC) at univariate step. At multivariate step, Cox's regression analysis was used to determine whether UBE2C IHC and Ki-67 IHC (11%) added independent prognostic information to NPI.
Sensitivity, specificity
Receiver operating characteristic (ROC) analysis with MR and death within 7 years as a defining point was computed. The area under the curve (AUC) was used as a measure of the marker global performance in the test set. ROC curves were calculated for UBE2C IHC and Ki-67 IHC (11%) and the bio-clinical prognostic index, NPI.
All data were analysed with SAS version 9.1 (SAS Institute, Cary, NC, USA) and STATA 10 SE (StataCorp, College Station, TX, USA).
Results
IHC
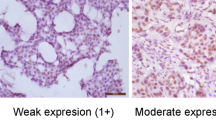
The UBE2C immunostaining was observed essentially in the nuclei of the carcinoma cells; however, in some cases, it was associated with a cytoplasmic immunostaining. The immunostaining was almost always of strong intensity (Figure 1A and B). For Ki-67 immunostaining, only nuclear staining was interpreted as positive; the intensity varied between mild and strong (Figure 1C and D). The patterns of UBE2C and Ki-67 immunostaining were similar. Exceptionally, for both Ki-67 and UBE2C, we observed nuclear immunostaining in rare normal ductal epithelial cells.
Immunohistochemistry staining of UBE2C and Ki-67 in node-positive breast carcinomas. (A) High expression of UBE2C with intense nuclear immunostaining of carcinoma cells. (B) Low expression of UBE2C with nuclear immunostaining of rare carcinoma cells. (C) High expression of Ki-67 with strong nuclear immunostaining of carcinoma cells. (D) Low expression of Ki-67 with rare immunostained carcinoma cells.
Statistical analysis
Descriptive analysis
The clinical and pathological characteristics of the 92 patients are detailed in Table 1. UBE2C IHC high level (⩾11%) was significantly related to other bad prognostic parameters such as high SBR grade, number of positive nodes, negative ER, high NPI and high Ki-67 IHC level (Table 2).
Inter-observer reproducibility
The UBE2C and Ki-67 IHC results are robust as they showed high inter-observer reproducibility (Supplementary Table 1 and Supplementary Figure 1). As continuous parameters, Spearman's Rho showed high correlation between both pathologists (DL and FL) for both parameters (ρ=0.977 – P<0.0001 and ρ=0.951 – P<0.0001 for UBE2C and Ki-67, respectively). When discretised (<11% vs ⩾11%), Kappa test also showed high agreement (κ=0.82 – P<0.0001 and κ=0.72 – P<0.0001 for UBE2C and Ki-67, respectively).
Correlation study between genomic and IHC data
Genomic and IHC data were available for 40 patients. Microarray values of the two UBE2C cDNA probes were significantly correlated with UBE2C IHC values (ρ=0.51 – P=0.0009 and ρ=0.65 – P<0.0001 for UMGC_2270 and UMGC_6429, respectively) (Supplementary Table 1 and Figure 2).
Correlation study between genomic and MFS
For the 40 patients with available UBE2C microarray data, prognostic value was confirmed for MFS prediction for both gene probes (hazard ratio (HR)=2.99, 95% confidence interval (CI))=(1.48–6.06) – P=0.002 and HR=2.21, 95% CI=(1.32–3.69) – P=0.002 for UMGC_2270 and UMGC_6429, respectively). Prognostic value was also confirmed for OS prediction for both UBE2C cDNA probes (HR=5.27, 95% CI=(2.36–11.74) – P<0.001 and HR=2.78, 95% CI=(1.62–4.76) – P<0.001 for UMGC_2270 and UMGC_6429, respectively). As UBE2C IHC and UBE2C genomic data were correlated, on the one hand, with each other and, on the other, with MFS, one can suggest the use of a more practicable technique (IHC) as a reliable way to measure the level of UBE2C and the transfer of this analysis to clinical routine.
Comparison of parameters for prognostic evaluation
Univariate step
UBE2C IHC and Ki-67 IHC were both significantly related to MFS and OS (Tables 3 and 4; Figures 3 and 4). Prognostic power for MFS, measured by AIC, was greater for NPI or UBE2C IHC than for Ki-67 IHC (199.62<202.89<208.74, respectively). Prognostic power for OS, measured by AIC, was greater for UBE2C IHC or NPI than for Ki-67 IHC (146.91<147.39<149.42, respectively). Moreover, ROC analysis (MR within 7 years as a defining point) showed that AUC was 0.77, 0.74 and 0.69 for NPI, UBE2C and Ki-67, respectively (P=0.17). For death (7 years as a defining point), AUC was 0.74, 0.69 and 0.66 for NPI, UBE2C and Ki-67, respectively (P=0.43). In conclusion, univariate survival prediction's performance of UBE2C IHC is close to that achieved by NPI, and both are better than Ki-67 IHC. The results of all bio-clinical parameters are listed in Tables 3 and 4.
Multivariate step
Multivariate analyses showed that, for MFS and OS, discretised (11%) UBE2C IHC (but not Ki-67 IHC) remained an independent factor that added prognostic information to bio-clinical index NPI, which was the most relevant bio-clinical parameter (Tables 5 and 6, data not shown).
Discussion
In this study, we have confirmed that increased expression of UBE2C protein was linked to poor prognosis in N+ breast cancer. Our result is in contradiction with Berlingieri et al (2007) who evaluated prognostic informativity of UBE2C and found no relation between this protein and the rates of overall and relapse-free survival. According to the following points, we strongly believe that UBE2C is a prognostic marker of breast cancer. First, we previously found the same result for this marker at the RNA level in a cohort of 252 node-positive breast cancer patients (Campone et al, 2008; Jézéquel et al, 2008). Second, UBE2C microarray data analysis of six other breast cancer genomic studies gave the same result in both N+ and N− patients (Jézéquel et al, 2008). Third, we found a significant correlation between UBE2C genomic values and UBE2C IHC values. Finally, our IHC study included more patients than did that of Berlingieri (92 instead of 74).
In regard to our recent studies and other ‘Omics’ studies, numerous components of the UPS have been found to be related to breast carcinogenesis and an unfavourable evolution in breast cancer (Chen and Madura, 2005; Ricolleau et al, 2006; Deng et al, 2007; Campone et al, 2008; Jézéquel et al, 2008). Furthermore, increased activity of the proteasome was directly linked to overexpression of UPS elements (ubiquitin enzymes and proteasomal subunits) (Chen and Madura, 2005). We can therefore hypothesise, first, that poor prognosis in N+ breast cancer is related in a large part to a high activity of the UPS, itself related to tumour high proliferative metabolism, and second, that UBE2C might be considered as a marker of proteasome activity.
On account of the central role of proteasome in protein homoeostasis and fate, whose dysregulation may lead to cancer, therapeutic strategies focused on this potential target; proteasome inhibitors could provide a new and promising class of anticancer agents (Ciechanover, 2003; Orlowsky and Dees, 2003; Sato et al, 2008). Since the discovery of bortezomib as proteasome inhibitor, this macromolecular protein assembly is to be considered as a therapeutic target. This drug has been approved by the Food and Drug Administration for treatment of relapsed and refractory multiple myeloma. But no effect was found in breast cancer when it was used as a single agent despite a proven efficacy when combined with several chemotherapeutic agents (Cusak, 2003). Two considerations may be advanced to explain this treatment failure. First, following the example of antioestrogens and hormone receptors, Herceptin and the HER2/neu receptor, it seems probable that the activity of the proteasome might be evaluated before any treatment with bortezomib (Marx et al, 2007). We propose that proteasome activity measurement could be determined by an indirect method: UBE2C IHC. To this end, we will soon compare a direct method by means of a fluorigenic substrate to UBE2C IHC (Chen and Madura, 2005). Second, according to recent study and the disappointing results of clinical trials using bortezomib as monotherapy in some solid tumours, it appears that this molecule should be used in combination. In 2006, Cardoso et al (2006) showed a synergy between bortezomib and trastuzumab in HER2/neu+++/++ cell lines. These results convinced the authors to conduct a phase 1 clinical trial that aimed at evaluating this drug combination.
On the basis of genomic data of a previous study, the hierarchical cluster analysis dendrogram of the 219 genes with the highest prognostic information in 252 N+ breast cancer patients showed a direct link between 2 proteasome subunit genes located in 17q12 and ERBB2 locus, known to be a hot spot of gene amplification in breast cancer. This cluster contains: PSMD3, PSMB3, STARD3, C17orf37 and ERBB2 (Supplementary Figure 2) (Jézéquel et al, 2008). Furthermore, PSMD3 and PSMB3 have been found to be overexpressed through gene amplification of ERBB2 locus in numerous studies (Kauraniemi et al, 2003; Buness et al, 2007). The relation between ERBB2 locus amplification, proteasome subunits overexpression and proteasome activity still needs to be explored, but the results exposed above may let us think that a functional link exists, and so a therapeutic approach should strongly consider this possibility.
In conclusion, in breast cancer, any new clinical trial testing bortezomib should compare bortezomib in combination (e.g., trastuzumab) vs the single molecule, and should include patients with proteasome high activity, which could be indirectly evaluated by UBE2C IHC staining.
Accession codes
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Berlingieri MT, Pallante P, Sboner P, Barbareschi M, Bianco M, Ferraro A, Mansueto G, Borbone E, Guerriero E, Troncone G, Fusco A (2007) UbcH10 is overexpressed in malignant breast carcinomas. Eur J Cancer 43: 2729–2735
Buness A, Kuner R, Ruschhaupt M, Poustka A, Sültmann H, Tresch A (2007) Identification of aberrant chromosomal regions from gene expression microarray studies applied to human breast cancer. Bioinformatics 23: 2273–2280
Campone M, Campion L, Roché H, Gouraud W, Charbonnel C, Magrangeas F, Minvielle S, Genève J, Martin AL, Bataille R, Jézéquel P (2008) Prediction of metastatic relapse in node-positive breast cancer: establishment of a clinicogenomic model after FEC100 adjuvant regimen. Breast cancer Res and Treat 109: 491–501
Cardoso F, Durbecq V, Laes JF, Badran B, Lagneaux L, Bex F, Desmedt C, Willard-Gallo K, Ross JS, Burny A, Piccart M, Sotiriou C (2006) Bortezomib (PS-341, Velcade) increases the efficacy of trastuzumab (Herceptin) in HER-2-positive breast cancer cells in a synergistic manner. Mol Cancer Ther 5: 3042–3051
Chen L, Madura K (2005) Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res 65: 5599–5606
Ciechanover A (2003) The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochem Soc Trans 31: 474–481
Cusak J (2003) Rationale for the treatment of solid tumors with the proteasome inhibitor bortezomib. Cancer Treat Rev 29: 21–31
Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, Dong L, Tang E, Yang H (2007) Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res and Treat 104: 21–30
Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C (2008) Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res 14: 5158–5165
Hershko A, Ciechanover A, Varshavsky A (2000) The ubiquitin system. Nature Med 6: 1073–1081
Jézéquel P, Campone M, Roché H, Gouraud W, Charbonnel C, Ricolleau G, Magrangeas F, Minvielle S, Genève J, Martin AL, Bataille R, Campion L (2008) 38-gene expression signature to predict metastasis risk in node-positive breast cancer after systemic adjuvant chemotherapy: a genomic substudy of PACS01 clinical trial. Breast Cancer Res and Treat (e-pub ahead of print; 27 May 2009)
Kauraniemi P, Kuukasjärvi T, Sauter G, Kallionemi A (2003) Amplification of a 280-kilobase core region at the ERBB2 locus leads to activation of two hypothetical proteins in breast cancer. Am J Pathol 163: 1979–1984
Lipkowitz S (2002) The role of the ubiquitination-proteasome pathway in breast cancer. Ubiquitin mediated degradation of growth factor receptors in the pathogenesis and treatment of cancer. Breat Cancer Res 5: 8–15
Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC (2003) Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA 100: 5974–5979
Mani A, Gelman E (2005) The ubiquitin-proteasome pathway and its role in Cancer. J Clin Oncol 21: 4776–4789
Marx C, Yau C, Banwait S, Zhou Y, Scott GK, Hann B, Park JW, Benz CC (2007) Proteasome-regulated ERBB2 and estrogen receptor pathways in breast cancer. Mol Pharmacol 6: 1525–1534
Ohta T, Fukuda M (2004) Ubiquitin and breast cancer. Oncogene 23: 2079–2088
Okamoto Y, Ozaki T, Miyazaki K, Aoyama M, Miyazaki M, Nakagawara A (2003) UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res 6: 4167–4173
Orlowsky RZ, Dees EC (2003) The role of the ubiquitination-proteasome pathway in breast cancer. Applying drugs that affect the ubiquitin-proteasome pathway to the therapy of breast cancer. Breast Cancer Res 5: 1–7
Ricolleau G, Charbonnel C, Lode L, Loussouarn D, Joalland MP, Bogumil R, Jourdain S, Minvielle S, Campone M, Déporte-Fety R, Campion L, Jézéquel P (2006) Surface-enhanced laser desorption/ionization time of flight mass spectrometry protein profiling identifies ubiquitin and ferritin light chain as prognostic biomarkers in node-negative breast cancer tumors. Proteomics 6.06: 1963–1975
Sato K, Rajendra E, Ohta T (2008) The UPS: a promising target for breast cancer treatment. BMC Biochem 9: 1–8
Stuart-Harris R, Caldas C, Pinder SE, Pharoah P (2008) Proliferation markers and survival in early breast cancer: a systematic review and met-analysis of 85 studies in 32,825 patients. Breast J 17: 323–334
Tanaka K (2009) The proteasome: overview of structure and functions. Proc Jpn Acad 85: 12–36
Viale G, Giobbe-Hurder A, Regan MM, Coates A, Mastropasqua MG, Dell’Orto P, Maiorano E, MacGrogan G, Braye SG, hlschlegel C, Neven P, Orosz Z, Olszewski WP, Knox F, Thürlimann B, Price KN, Castiglione-Gertsch M, Gelber RD, Gusterson BA, Goldhirsch A (2008) Prognostic and predictive value of centrally reviewed Ki-67 labeling index in postmenopausal women with endocrine-responsive breast cancer: results from breast international group trial 1–98 comparing adjuvant tamoxifen with letrozole. J Clin Oncol 26: 5569–5575
Wagner KW, Sapinoso LM, El-Rifai W, Frierson HF, Butz N, Mestan J, Hofmann F, Deveraux QL, Hampton GM (2004) Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene 23: 6621–6629
Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schütz F, Goldstein DR, Piccart M, Delorenzi M (2008) Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res 10: R65
Acknowledgements
This study was supported by SANOFI-AVENTIS, France and PFIZER, France. We thank E Biraud and V Véron for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Loussouarn, D., Campion, L., Leclair, F. et al. Validation of UBE2C protein as a prognostic marker in node-positive breast cancer. Br J Cancer 101, 166–173 (2009). https://doi.org/10.1038/sj.bjc.6605122
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605122
Keywords
This article is cited by
-
ClearF: a supervised feature scoring method to find biomarkers using class-wise embedding and reconstruction
BMC Medical Genomics (2019)
-
Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer
Cellular & Molecular Biology Letters (2019)
-
The clinicopathological significance of UBE2C in breast cancer: a study based on immunohistochemistry, microarray and RNA-sequencing data
Cancer Cell International (2017)
-
Ubiquitin-conjugating enzyme E2C regulates apoptosis-dependent tumor progression of non-small cell lung cancer via ERK pathway
Medical Oncology (2015)
-
Prognostic value of different cut-off levels of Ki-67 in breast cancer: a systematic review and meta-analysis of 64,196 patients
Breast Cancer Research and Treatment (2015)