Abstract
The epidermal growth factor receptor (EGFR) family plays an important role in breast carcinogenesis. Much interest has been focused recently on its members because of their potential role as prognostic indicators in breast cancer and their involvement in cancer therapy. We have evaluated more than 1500 cases of invasive breast carcinoma immunohistochemically using tissue microarray technology to examine the expression of EGFR family receptor proteins. We have found that 20.1 and 31.8% of cases were positive for EGFR and c-erbB-2, respectively, and 45 and 45.1% of tumours overexpressed for c-erbB-3 and c-erbB-4, respectively. The expression of either EGFR or c-erbB-2 was associated with other bad prognostic features and with poor outcome. Neither c-erbB-3 nor c-erbB-4 had any association with survival. c-erbB-2 had an independent prognostic effect on overall and disease-free survival (DFS) in all cases, as well as in the subset of breast carcinoma patients with nodal metastases. Several hetero- and homodimeric combinations have been reported between the EGFR members. Those dimers can evoke diverse signal transduction pathways with variable cellular responses. We stratified cases according to their co-expression of receptors into distinct groups with different receptor-positive combinations. Patients whose tumours co-expressed c-erbB-2 and c-erbB-3, as well as those whose tumours co-expressed EGFR, c-erbB-2 and c-erbB-4 showed an unfavourable outcome compared with other groups, while combined c-erbB-3 and c-erbB-4 expression was associated with a better outcome. In cases showing expression of one family member only (homodimers), we found a significant association between c-erbB-4 homodimer-expressing tumours and better DFS. In contrast, patients with c-erbB-2 homodimer-expressing tumours had a significant poorer DFS compared with other cases. These data imply that the combined profile expression patterns of the four receptor family members together provide more accurate information on the tumour behaviour than studying the expression of each receptor individually.
Similar content being viewed by others
Main
The epidermal growth factor receptor (EGFR) tyrosine kinase family consists of four members: EGFR, c-erbB-2, c-erbB-3 and c-erbB-4 (Gullick and Srinivasan, 1998). All share structural homology consisting of an extracellular ligand-binding domain, a transmembrane domain and an intracytoplasmic tyrosine kinase domain (Mason and Gullick, 1995). There is a growing body of evidence that this family is involved in breast cancer development and progression (Gullick and Srinivasan, 1998). Previous studies of EGFR and c-erbB-2 reported their association with characteristics of poor prognosis. Epidermal growth factor receptor overexpression was always associated with poor outcome, manifested in a short overall survival and disease-free interval (Sainsbury et al, 1985; Lewis et al, 1990; Toi et al, 1994; Tsutsui et al, 2002), high metastatic potential (Sainsbury et al, 1987) and negative oestrogen receptor (ER) status (Pilichowska et al, 1997; Tsutsui et al, 2002). The same behaviour was also detected in tumours showing c-erbB-2 gene amplification and/or overexpression, which was marked in high tumour grade as well as short overall survival and disease-free interval (Winstanley et al, 1991; Charpin et al, 1997; Suo et al, 2002).
Relatively less information has been reported about the other two members: c-erbB-3 and c-erbB-4. Significant correlations have been found between c-erbB-3 overexpression and tumour size (Travis et al, 1996) and histological grade (Naidu et al, 1998). Studies on c-erbB-4 expression have shown an association with well-differentiated tumours (Kew et al, 2000; Suo et al, 2002) and positive ER status (Suo et al, 2001). Some studies have found an association with better survival (Pawlowski et al, 2000; Suo et al, 2002), while others have reported an association with poor survival (Lodge et al, 2003) or no association with survival (Kew et al, 2000).
Most of the previous research has focused on investigating the expression of individual members in relation to different clinicopathological studies. However, a few studies have shown that the co-expression of two or more members had an adverse effect on breast cancer behaviour and outcome. The best example for these findings is the reported adverse synergistic effect of EGFR and c-erbB-2 expression on both prognosis (Osaki et al, 1992; Toi et al, 1994; Suo et al, 2002) and metastasis (Brandt et al, 1999). Such studies have evoked our interest to investigate the expression of the four members together on a large number of invasive breast carcinomas with long follow-up in order to shed light on any potential prognostic implications.
Materials and methods
Patients
A consecutive series of 1944 cases of primary operable invasive breast carcinoma from patients presenting between 1986 and 1998 and entered into the Nottingham Tenovus Primary Breast Carcinoma Series were used. Data on histological grade (Elston and Ellis, 1991), histological tumour type (Ellis et al, 1992), vascular invasion (Pinder et al, 1994), tumour size, lymph node stage and Nottingham Prognostic Index (NPI) (Galea et al, 1992) are routinely assessed and recorded in the database. The NPI is calculated using the following equation: NPI=0.2 × tumour size (cm)+grade (1–3)+lymph node stage (1–3). Patients are assigned into three groups: good, moderate and poor. A score of ⩽3.4 indicates a good prognosis, 3.41–5.4 a moderate prognosis and >5.4 a poor prognosis.
Patient age ranged from 18 to 70 years (mean age 53, median 54 years). Mean survival was 62 months (range 1–192 months). Information on local, regional and distant recurrence and survival is maintained on a prospective basis. Patients are followed up at 3-month intervals initially, then 6 monthly and then annually, for a median period of 58 months. The disease-free interval was defined as the interval (in months) from the date of the primary surgical treatment to the first loco-regional or distant recurrence. The overall survival was taken as the time (in months) from the date of the primary surgical treatment to the time of death. Oestrogen receptor status was estimated immunohistochemically in 1805 of the tumours; 553 (30.6%) were negative for ER expression, while 1252 (69.4%) carcinomas were ER positive.
Data for histological tumour type, grade and staging are summarised in Tables 1 and 2.
Construction of the tissue microarray blocks
Breast cancer tissue microarrays were prepared as described previously (Kononen et al, 1998; Camp et al, 2000; Torhorst et al, 2001). Haematoxylin and eosin (H&E) slides were obtained from each available conventional tumour block and used as a guide for selection of the most representative areas of the tumour. Tissue microarrays were constructed by obtaining 0.6 mm diameter cylinders from the original blocks and re-embedding these cores into the recipient block. Each case was sampled twice, from the centre and the periphery of the tumour, to form an array of 100 cases per block. Histological tumour types and tumour grade are summarised in Tables 1 and 2.
Immunohistochemistry
Immunohistochemical staining for the sections was performed according to the avidin–biotin complex method. Tissue sections of 3 μm thickness were taken from tissue array blocks. The initial sections were stained with H&E to confirm the histological diagnosis. Paraffin sections were dewaxed and then rehydrated. To block the endogenous peroxidase, the rehydrated sections were treated with 0.3% hydrogen peroxide in methanol for 10 min. To unmask the antigens, sections, with the exception of those for c-erbB-2 and c-erbB-4, were microwaved in citrate buffer, pH 6 for a total 20 min. After the nonspecific staining had been blocked by normal swine serum, sections were incubated with the primary antibodies for between 50 and 60 min. The antibodies used were EGFR (clone EGFR.113, Novocastra, diluted at 1 : 10), c-erbB-2 (Dako, diluted at 1 : 250), c-erbB-3 (clone RTJ1, Novocastra, diluted at 1 : 20), c-erbB-4 (clone HFR1, Neomarkers, diluted at 6 : 4) and ER (clone 1D5, Dako, diluted at 1 : 80). Sections were incubated with the biotin-labelled secondary antibody (diluted 1 : 100) for 30 min, then in avidin–biotin complex (diluted 1 : 100) for a further 45 min. 3-3′Diaminobenzidine tetrahydrochloride was used as the chromogen.
Controls
Positive and negative controls were included in each staining run. Positive controls were myoepithelial cells of normal duct in normal mammary gland for EGFR, renal tissue (proximal and distal tubules) for c-erbB-3 and known positive cases of breast carcinoma for c-erbB-2, c-erbB-4 and ER. Negative controls were obtained by omitting the primary antibodies.
Immunohistochemical scoring
The modified histochemical score (H-score) (McCarty et al, 1985) was used as it includes a semiquantitative assessment of both the intensity of staining and the percentage of positive cells. For the intensity, a score of 0–3 corresponding to negative, weak, moderate and strong positivity was recorded. In addition, the percentage of positive cells at each intensity was estimated in %. The H-score is calculated as (1 × weak %+2 × moderate %+3 × strongly stained %). The range of possible scores is thus 0–300.
Two cores were evaluated from each tumour. Each core was scored individually, then the mean of the two readings was calculated. Only the invasive carcinoma was assessed for staining. Noninvasive cells such as stromal cells, normal epithelial cells, benign lesions and carcinoma in situ were excluded from assessment. If one core was uninformative (either lost or contained no tumour tissues), the overall score applied was that of the remaining core. Previous studies have validated the use of one core to study the expression of tumour markers having heterogeneous distribution (Camp et al, 2000; Torhorst et al, 2001). One observer scored all cases, which were re-checked randomly by the same investigator after a period of time. A good correlation was found between the two estimations.
The cutoff points of expression were determined according to frequency histograms. For all markers, tumours with 5% of the neoplastic cells showing immunoreactivity were considered positive, while those with less than 5% were classified negative. Additionally, for c-ebB-3 and c-erbB-4, we considered the median as a cutoff between weak and strong expression (150 and 100, respectively). For ER, the cutoff point was taken at an H-score of 20.
Statistical analysis
Association between the immunohistochemical findings and different clinicopathological parameters was evaluated by χ2 test. A P-value of <0.05 was considered to reflect a significant relationship. Survival curves were calculated by the Kaplan–Meier method. The differences between survivals were estimated by log-rank test. Multivariate Cox regression analysis was used to evaluate whether there was any independent prognostic effect of the variables on disease-free interval or overall survival.
Results
Epidermal growth factor receptor family protein expression (Figure 1)
Epidermal growth factor receptor protein expression was studied in 1584 cases. The expression was mainly cytoplasmic and membranous and identified in 20.1% of cases. The expression of c-erbB-2 protein was investigated in 1812 breast cancers, of which 31.8% showed membranous immunoreactivity. For the 1499 cases stained with c-erbB-3, expression was mainly cytoplasmic, with only 10.7% of cases considered negative, 44.3% weakly positive and 45% strongly positive. In the 1513 cases stained for c-erbB-4, predominantly cytoplasmic reactivity was identified in 79.3% of the tumours (34.2% weak and 45.1% strong positive), while 20.7% were negative.
Significant positive associations were found between all EGFR family members (Table 3), where 37.3% of HER1-positive tumours are HER2 positive and 24.3% of HER2-positive cases are positive for HER1 (χ2=7.01, P=0.008); 54.7% of HER1-positive cases are overexpressing to HER3 and 23.9% of HER3-overexpressing tumours are HER1 positive (χ2=14.54, P=0.001); 55.8% of HER1-positive tumours are overexpressing to HER4 and 24.3% of HER4 overexpressing cancers are positive to HER1 (χ2=17.60, P<0.001); 50.1% of HER2-positive tumours are overexpressing to HER3 and 34.9% of HER3-overexpressing cases are HER2 positive (χ2=8.66, P=0.013); 49.5% of HER2-positive tumours are overexpressing to HER4 and 34.6% of HER4-overexpressing tumours are positive to HER2 (χ2=6.41, P=0.040); 64.9% of HER3-overexpressing tumours are also overexpressing for HER4 and 63.7% of HER4-overexpressing tumours are also overexpressing for HER3 (χ2=351.90, P<0.001).
Association between EGFR family members and different clinicopathological parameters
Table 4 shows the results of statistical analyses of the correlations between different members and clinical and pathological variables. Epidermal growth factor receptor expression was significantly associated with higher tumour grade, increasing size, higher NPI, the development of distant metastases and the incidence of death, but inversely correlated with ER status. Regarding c-erbB-2, immunoreactivity was significantly associated with poorer grade, higher NPI, local and regional recurrence, distant metastases and death, while inversely correlated to ER status. The expression of c-erbB-3 showed a significant inverse association with local recurrence, although it was significantly associated with poorer grade, lymph node disease, higher NPI and the presence of vascular invasion. The c-erbB-4 receptor expression showed a significant paradoxical association with ER status and local recurrence, while being significantly associated with poorer histological grade, higher lymph node stage, NPI and the presence of vascular invasion.
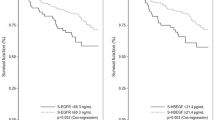
Survival analyses were performed comparing the expression of the four markers in relation to both DFS and OS. We found that EGFR expression was significantly associated with shorter disease-free survival (DFS; P=0.0265) and overall survival (P=0.0035). c-erbB-2 was also significantly correlated with poorer overall survival (P=0.0006) and DFS (P=0.0001) (Figure 2A and B). Neither c-erbB-3 nor c-erbB-4 showed a significant association with DFS (P=0.3401 and 0.2555, respectively) or overall survival (P=0.7462 and 0.7747, respectively).
(A) Epidermal growth factor receptor expression in relation to overall survival demonstrating survival differences between negative and positive cases. (B) Epidermal growth factor receptor expression in relation to DFS showing longer DFS in negative patients. (C) c-erbB-2 in relation to overall survival demonstrating survival differences between negative and positive cases. (D) c-erbB-2 in relation to DFS with better DFS in c-erbB-2-negative cases.
Cox multivariate analyses showed that c-erbB-2 was not only significantly related to DFS and overall survival, but also that its predictive power was independent of histological grade, tumour size, lymph node status and ER status in all patients as well as patients with nodal spread. In node-negative patients, it had no prognostic power in predicting OS, while its prognostic impact is preserved in predicting DFS (Tables 5 and 6).
Co-expression of EGFR family members and their association with OS and DFS
To define the frequency of dimers formation, we combined the expression of the four markers in the 1406 cases of breast cancer in which the results were available for all four members of the EGFR family. In this part of the analysis, c-erbB-3- and c-erbB-4-overexpressing cases were considered to be those showing immunohistochemical positivity above the median level, while a level below the median was considered as normal expression.
The frequencies and percentage of different dimers are shown in Table 7.
The Kaplan–Meier estimates (Table 8) for the groups of combined co-expression of two or more receptors and both of overall and relapse-free survival showed a significant positive correlation between combined c-erbB-2 and c-erbB-3 expression and reduced OS (Figure 3A and B). The same relationship was also noticed with combined expression of EGFR, c-erbB-2 and c-erbB-4 in relation to OS and DFS (Figures 4A and B) and for c-erbB-2 homodimers in relation to DFS (Figure 6) only. Conversely, the combined expression of c-erbB-3 and c-erbB-4 was significantly associated with a better OS and DFS (Figure 5A and B). Also, c-erbB-4 homodimer was significantly associated with a better DFS (Figure 7).
We were interested to see whether there was a significant difference between c-erbB-2/c-erbB-3 heterodimer expression and c-erbB-2/c-erbB-2 homodimer expression in relation to DSF and OS. Although no significant differences were detected between these two subgroups in relation to DFS (P=0.6606) and OS (P=0.2501), we found that OS was worse in patients with tumours expressing c-erbB-2/c-erbB-3 heterodimers compared to those with tumours expressing c-erbB-2/c-erbB-2 homodimer (Figure 8).
Discussion
In this study, expression of EGFR was identified in 20.1% of cases, consistent with previous studies where EGFR expression has been reported in 14–65% of breast cancer (Suo et al, 1998; Walker and Dearing, 1999). c-erbB-2 overexpression was detected in 31.8%, in line with previous reports showing 10–34% expression (Pauletti et al, 1996; Ross and Fletcher, 1999). Expression of c-erbB-3 and c-erbB-4 was 89.3% (45% strongly positive) and 79.3% (45.1% strongly positive), respectively. These findings are also in accordance with previous work which has identified c-erbB-3 expression in 65–95% of cases and c-erbB-4 in 58–82% of breast cancers (Travis et al, 1996; Naidu et al, 1998; Kew et al, 2000; Suo et al, 2002; Witton et al, 2003).
In this study, EGFR expression was significantly associated with features recognised to reflect poor prognosis, including high (poor) histological grade, high NPI score, negative ER status, larger tumour size, the development of distant metastases and death. Previous studies have reported the same relationships with tumour grade (Suo et al, 1998), negative ER status and reduced survival (Toi et al, 1990). Other studies have failed to find an association with grade, tumour size and lymph node status (Lewis et al, 1990).
We have confirmed the previously reported significant correlation identified between c-erbB-2-expressing tumours and poorer tumour grade, ER negativity (Lovekin et al, 1991; Suo et al, 2002; Witton et al, 2003; Zhang et al, 2003), high NPI score, local and regional recurrence, distant metastases and death from breast cancer. Our finding regarding the positive association between the development of distant metastatic disease and c-erbB-2 expression corroborates the findings of Tan et al (1997), who transfected c-erbB-2 gene in the very low c-erbB-2-expressing MDA-MB-435 human breast cancer cell line and found that its overexpression enhanced the metastatic potential of cancer cells.
A significant association between c-erbB-2 overexpression and poor outcome in the form of short OS and DFS was found, as previously observed in several series (Lovekin et al, 1991; Winstanley et al, 1991; Witton et al, 2003). Winstanley et al (1991) showed the independent prognostic effect in relation to OS. We have also demonstrated that c-erbB-2 expression independently predicted for poor OS and DFS in the whole population of breast cancer patients as well as in those with node-positive disease. The significant impact of c-erbB-2 overexpression/amplification on poor outcome in patients with positive nodal metastases is well established (Lovekin et al, 1991; O'Reilly et al, 1991).
In this series, c-erbB-3 expression was positively associated with grade, lymph node metastases, poor NPI and vascular invasion, while it was inversely associated with local recurrence. No relation was identified with either ER status or survival. Previous reports have recorded an association between c-erbB-3 overexpression and poor prognostic features such as high grade (Naidu et al, 1998) and lymph node spread (Lemoine et al, 1992). Some other studies have previously noted an association with ER status, which we did not find in the present large series. For example, Knowlden et al (1998) reported a significant association with positive ER status and good response to endocrine therapy for cases overexpressing c-erbB-3, while others have failed to find associations with ER status (Travis et al, 1996; Naidu et al, 1998). We have also, in the present study, identified an inverse relation with local recurrence, while the reverse was reported by Travis et al (1996), where they found that moderate to strong expressing tumours were more likely to develop local recurrence compared with weak expressing and negative tumours (Travis et al, 1996). The discrepancies in such relations may be partly explained by the discovery of a c-erbB-3-soluble form, p85-soluble ErbB-3 (sErbB-3), a potent negative regulator to heregulin, which inhibits heregulin-induced phosphorylation of c-erbB-2, c-erbB-3 and c-erbB-4 by competing for heregulin binding (Lee et al, 2001), although other potential explanations may also be variations in antibodies and cutoffs used.
Very little is known with respect to the prognostic significance of c-erbB-4 in breast cancer. In this study, we have demonstrated that its overexpression was associated with higher grade, nodal metastases, poor NPI and vascular invasion, and inversely correlated with ER status and local recurrence. No significant association was identified in relation to survival. Most studies presented to date do not have congruent findings regarding the prognostic effect of c-erbB-4 in breast cancer. Some studies linked c-erbB4 overexpression with a well-differentiated phenotype (Kew et al, 2000; Suo et al, 2002), positive ER status (Knowlden et al, 1998; Suo et al, 2001) and longer survival (Suo et al, 2002; Witton et al, 2003). Another study associated overexpression with poor survival in lymph node-positive cases (Lodge et al, 2003). The significant inverse association between c-erbB-4 expression and ER status observed in the present study has only been recorded in one study by Vogt et al (1998), who demonstrated an inverse association between c-erbB-4 gene amplification and ER status in breast cancer (Vogt et al, 1998). These conflicting results may be related to different scoring systems used, different definitions of overexpression, the use of different antibodies, antigen retrieval techniques and the heterogeneous populations of patients selected for the studies. Variable responses may also be evoked by signals generated from either c-erbB-4 homodimers or heterodimers with other members, the level of which depends upon the context of expression of other receptors. This may explain the association of c-erbB-4 overexpression with some poor prognostic features in our study, as a great many of cases co-expressed one or more other receptors along with c-erbB-4. In support of this concept, a poor prognostic association has been reported in relation to the co-expression of c-erbB-2 and c-erbB-4 in childhood medulloblastoma (Gilbertson et al, 1997). Another important factor in relation to the diverse behaviour of c-erbB-4 is the type of isoform expressed by the tumour. For example, the CYT-1 isoform mediates proliferation as well as chemotaxis and survival signals, whereas the CYT-2 isoform stimulates proliferation and growth only (Junttila et al, 2000).
The hallmark of this family of growth factor receptors is the ability of its members to act or function synergistically with another receptor through dimerisation. ErbB-2 represents the preferred heterodimerisation partner of all other receptors of the family, and the preferred dimerisation partner of ErbB-2 is ErbB-3 (Tzahar et al, 1996). We found that 50.1% of c-erbB-2-expressing tumours also overexpressed c-erbB-3 and that this heterodimerisation was significantly associated with poor OS and DFS. This dimer, formed of ligand-deficient c-erbB-2 and kinase-deficient c-erbB-3, is known to form the most potent signalling pair in terms of growth and transformation (Alimandi et al, 1995; Wallasch et al, 1995). Therefore, the detection of the co-expression of these two receptors may have more clinical and prognostic significance than the detection of expression of each receptor separately.
Although there was a significant association between EGFR and c-erbB-2 expression in this study, dimers formed of these receptors alone are less frequent than other dimers; however, their combined expression was more frequently common with c-erbB-3 and c-erbB-4. Our study was one of few studies that addressed the expression of the four receptors together compared to previous studies that considered the expression of EGFR and c-erbB-2 ignoring the other receptors (Tsutsui et al, 2003).
In spite of the fact that the immunohistochemical expression of both of EGFR and c-erbB-2 are linked with poor survival, heterodimers formed of this pair had no significant association with survival. It was surprising that EGFR and c-erbB-2 alone had prognostic impacts whereas the combination of both was of no significance, which disagree with literature (Toi et al, 1994; Suo et al, 2002). We have no explanation for this contradiction, but there are several factors that potentially contribute to these inconsistent findings, including the use of different antibodies and different cutoffs and definition of overexpression. Changing the antibodies against EGFR and/or c-erbB-2 might overcome this contradiction.
Cases co-expressing more than two family members, the combination of EGFR, c-erbB-2 and c-erbB-4 had a highly significant worse OS and DFS compared to other cases. The co-expression of EGFR and c-erbB-2 has an additive adverse effect in relation to survival (Toi et al, 1994; Suo et al, 2002).
Another interesting heterodimer identified in our study is that of c-erbB-3 and c-erbB-4; cases which expressed both receptors had a significantly better OS and DFS (trend) compared with other dimers. We have no explanation for this finding; however, cell line studies have shown that NDF induced mitogenesis in cells expressing c-erbB-3 or c-erbB-4 but not transformation, which was induced only when either EGFR or c-erbB-2 were coexpressed with c-erbB-3 or c-erbB-4 (Zhang et al, 1996). This suggests that c-erbB-3 and c-erbB-4 may activate signalling pathways that are different from those activated by EGFR and c-erbB-2, or an additional pathway may be needed to induce transformation.
In the present study, c-erbB-2 homodimers were significantly associated with a poorer DFS. In spite of being a ligandless receptor, cell line studies have shown that its overexpression induced homodimerisation that was sufficient to induce growth, malignant transformation (Di Fiore et al, 1987; Brennan et al, 2000) and cell migration (Verbeek et al, 1998). Conversely, we found that c-erbB-4 homodimers were significantly associated with a better DFS compared with others. A previous study has similarly recorded an association with a differentiated phenotype and with better prognosis in breast cancer (Suo et al, 2002), and another recent study has reported that breast cancer cases overexpressing c-erbB-4 only were the best of all cases regarding outcome (Witton et al, 2003).
The diversity identified in our study, in terms of survival, between different heterodimers and homodimers may be useful in subgrouping of breast cancer patients with significantly differing outcome. These data suggest that the predictive value of EGFR family overexpression may be optimised by combining information about the expression of all of the family members, rather than the assessment of a single receptor in isolation.
There are other essential components of the EGFR family network, which have not been studied here; 10 or more ligands and the proteins that are involved in EGFR family-induced pathways. The receptors are conduits for the ligand-activated signalling pathways and, although their expression levels clearly do contain useful information, the precision of the analysis can only be improved in the future by using antibodies for detection of such proteins.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Di Fiore PP, Kraus MH (1995) Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene 10: 1813–1821
Brandt BH, Roetger A, Dittmar T, Nikolai G, Seeling M, Merschjann A, Nofer JR, Dehmer-Moller G, Junker R, Assmann G, Zaenker KS (1999) c-erbB-2/EGFR as dominant heterodimerization partners determine a motogenic phenotype in human breast cancer cells. FASEB J 13: 1939–1949
Brennan PJ, Kumagai T, Berezov A, Murali R, Greene MI, Kumogai T (2000) HER2/neu: mechanisms of dimerization/oligomerization. Oncogene 19: 6093–6101
Camp RL, Charette LA, Rimm DL (2000) Validation of tissue microarray technology in breast carcinoma. Lab Invest 80: 1943–1949
Charpin C, Garcia S, Bouvier C, Martini F, Lavaut MN, Allasia C, Bonnier P, Andrac L (1997) c-erbB-2 oncoprotein detected by automated quantitative immunocytochemistry in breast carcinomas correlates with patients' overall and disease-free survival. Br J Cancer 75: 1667–1673
Di Fiore PP, Pierce JH, Kraus MH, Segatto O, King CR, Aaronson SA (1987) erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science 237: 178–182
Ellis IO, Galea M, Broughton N, Locker A, Blamey RW, Elston CW (1992) Pathological prognostic factors in breast cancer. II. Histological type. Relationship with survival in a large study with long-term follow-up. Histopathology 20: 479–489
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19: 403–410
Galea MH, Blamey RW, Elston CE, Ellis IO (1992) The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 22: 207–219
Gilbertson RJ, Perry RH, Kelly PJ, Pearson AD, Lunec J (1997) Prognostic significance of HER2 and HER4 coexpression in childhood medullablastoma. Cancer Res 57: 3272–3280
Gullick WJ, Srinivasan R (1998) The type 1 growth factor receptor family: new ligands and receptors and their role in breast cancer. Breast Cancer Res Treat 52: 43–53
Junttila TT, Sundvall M, Maatta JA, Elenius K (2000) Erbb4 and its isoforms: selective regulation of growth factor responses by naturally occurring receptor variants. Trends Cardiovasc Med 10: 304–310
Kew TY, Bell JA, Pinder SE, Denley H, Srinivasan R, Gullick WJ, Nicholson RI, Blamey RW, Ellis IO (2000) c-erbB-4 protein expression in human breast cancer. Br J Cancer 82: 1163–1170
Knowlden JM, Gee JM, Seery LT, Farrow L, Gullick WJ, Ellis IO, Blamey RW, Robertson JF, Nicholson RI (1998) c-erbB3 and c-erbB4 expression is a feature of the endocrine responsive phenotype in clinical breast cancer. Oncogene 17: 1949–1957
Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4: 844–847
Lee H, Akita RW, Sliwkowski MX, Maihle NJ (2001) A naturally occurring secreted human ErbB3 receptor isoform inhibits heregulin-stimulated activation of ErbB2, ErbB3, and ErbB4. Cancer Res 61: 4467–4473
Lemoine NR, Barnes DM, Hollywood DP, Hughes CM, Smith P, Dublin E, Prigent SA, Gullick WJ, Hurst HC (1992) Expression of the ERBB3 gene product in breast cancer. Br J Cancer 66: 1116–1121
Lewis S, Locker A, Todd JH, Bell JA, Nicholson R, Elston CW, Blamey RW, Ellis IO (1990) Expression of epidermal growth factor receptor in breast carcinoma. J Clin Pathol 43: 385–389
Lodge AJ, Anderson JJ, Gullick WJ, Haugk B, Leonard RC, Angus B (2003) Type 1 growth factor receptor expression in node positive breast cancer: adverse prognostic significance of c-erbB-4. J Clin Pathol 56: 300–304
Lovekin C, Ellis IO, Locker A, Robertson JF, Bell J, Nicholson R, Gullick WJ, Elston CW, Blamey RW (1991) c-erbB-2 oncoprotein expression in primary and advanced breast cancer. Br J Cancer 63: 439–443
Mason S, Gullick WJ (1995) Type 1 growth factor receptors: an overview of recent developments. Breast 4: 11–18
McCarty Jr KS, Miller LS, Cox EB, Konrath J, McCarty Sr KS (1985) Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109: 716–721
Naidu R, Yadav M, Nair S, Kutty MK (1998) Expression of c-erbB3 protein in primary breast carcinomas. Br J Cancer 78: 1385–1390
O'Reilly SM, Barnes DM, Camplejohn RS, Bartkova J, Gregory WM, Richards MA (1991) The relationship between c-erbB-2 expression, S-phase fraction and prognosis in breast cancer. Br J Cancer 63: 444–446
Osaki A, Toi M, Yamada H, Kawami H, Kuroi K, Toge T (1992) Prognostic significance of co-expression of c-erbB-2 oncoprotein and epidermal growth factor receptor in breast cancer patients. Am J Surg 164: 323–326
Pauletti G, Godolphin W, Press MF, Slamon DJ (1996) Detection and quantitation of HER-2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene 13: 63–72
Pawlowski V, Revillion F, Hebbar M, Hornez L, Peyrat JP (2000) Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res 6: 4217–4225
Pilichowska M, Kimura N, Fujiwara H, Nagura H (1997) Immunohistochemical study of TGF-alpha, TGF-beta1, EGFR, and IGF-1 expression in human breast carcinoma. Mod Pathol 10: 969–975
Pinder SE, Ellis IO, Galea M, O'Rouke S, Blamey RW, Elston CW (1994) Pathological prognostic factors in breast cancer. III. Vascular invasion: relationship with recurrence and survival in a large study with long-term follow-up. Histopathology 24: 41–47
Ross JS, Fletcher JA (1999) HER-2/neu (c-erb-B2) gene and protein in breast cancer. Am J Clin Pathol 112: S53–S67
Sainsbury JR, Farndon JR, Needham GK, Malcolm AJ, Harris AL (1987) Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. Lancet 1: 1398–1402
Sainsbury JR, Malcolm AJ, Appleton DR, Farndon JR, Harris AL (1985) Presence of epidermal growth factor receptor as an indicator of poor prognosis in patients with breast cancer. J Clin Pathol 38: 1225–1228
Suo Z, Berner HS, Risberg B, Karlsson MG, Nesland JM (2001) Estrogen receptor-alpha and C-ERBB-4 expression in breast carcinomas. Virchows Arch 439: 62–69
Suo Z, Emilsen E, Tveit KM, Nesland JM (1998) Type 1 protein tyrosine kinases in benign and malignant breast lesions. Histopathology 33: 514–521
Suo Z, Risberg B, Kalsson MG, Willman K, Tierens A, Skovlund E, Nesland JM (2002) EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J Pathol 196: 17–25
Tan M, Yao J, Yu D (1997) Overexpression of the c-erbB-2 gene enhanced intrinsic metastasis potential in human breast cancer cells without increasing their transformation abilities. Cancer Res 57: 1199–1205
Toi M, Nakamura T, Mukaida H, Wada T, Osaki A, Yamada H, Toge T, Niimoto M, Hattori T (1990) Relationship between epidermal growth factor receptor status and various prognostic factors in human breast cancer. Cancer 65: 1980–1984
Toi M, Tominaga T, Osaki A, Toge T (1994) Role of epidermal growth factor receptor expression in primary breast cancer: results of a biochemical study and an immunocytochemical study. Breast Cancer Res Treat 29: 51–58
Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Kochli OR, Mross F, Dieterich H, Moch H, Mihatsch M, Kallioniemi OP, Sauter G (2001) Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 159: 2249–2256
Travis A, Pinder SE, Robertson JF, Bell JA, Wencyk P, Gullick WJ, Nicholson RI, Poller DN, Blamey RW, Elston CW, Ellis IO (1996) C-erbB-3 in human breast carcinoma: expression and relation to prognosis and established prognostic indicators. Br J Cancer 74: 229–233
Tsutsui S, Ohno S, Murakami S, Hachitanda Y, Oda S (2002) Prognostic value of epidermal growth factor receptor (EGFR) and its relationship to the estrogen receptor status in 1029 patients with breast cancer. Breast Cancer Res Treat 71: 67–75
Tsutsui S, Ohno S, Murakami S, Kataoka A, Kinoshita J, Hachitanda Y (2003) Prognostic value of the combination of epidermal growth factor receptor and c-erbB-2 in breast cancer. Surgery 133: 219–221
Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y (1996) A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol 16: 5276–5287
Verbeek BS, Adriaansen-Slot SS, Vroom TM, Beckers T, Rijksen G (1998) Overexpression of EGFR and c-erbB2 causes enhanced cell migration in human breast cancer cells and NIH3T3 fibroblasts. FEBS Lett 425: 145–150
Vogt U, Bielawski K, Schlotter CM, Bosse U, Falkiewicz B, Podhajska AJ (1998) Amplification of erbB-4 oncogene occurs less frequently than that of erbB-2 in primary human breast cancer. Gene 223: 375–380
Walker RA, Dearing SJ (1999) Expression of epidermal growth factor receptor mRNA and protein in primary breast carcinomas. Breast Cancer Res Treat 53: 167–176
Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A (1995) Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J 14: 4267–4275
Winstanley J, Cooke T, Murray GD, Platt-Higgins A, George WD, Holt S, Myskov M, Spedding A, Barraclough BR, Rudland PS (1991) The long term prognostic significance of c-erbB-2 in primary breast cancer. Br J Cancer 63: 447–450
Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM (2003) Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol 200: 290–297
Zhang D, Salto-Tellez M, Putti TC, Do E, Koay ES (2003) Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod Pathol 16: 79–84
Zhang K, Sun J, Liu N, Wen D, Chang D, Thomason A, Yoshinaga SK (1996) Transformation of NIH 3T3 cells by HER3 or HER4 receptors requires the presence of HER1 or HER2. J Biol Chem 271: 3884–3890
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Abd El-Rehim, D., Pinder, S., Paish, C. et al. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer 91, 1532–1542 (2004). https://doi.org/10.1038/sj.bjc.6602184
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6602184
Keywords
This article is cited by
-
The clinical significance of cyclin B1 (CCNB1) in invasive breast cancer with emphasis on its contribution to lymphovascular invasion development
Breast Cancer Research and Treatment (2023)
-
Domain-level epitope mapping of polyclonal antibodies against HER-1 and HER-2 receptors using phage display technology
Scientific Reports (2022)
-
The role of HER2 alterations in clinicopathological and molecular characteristics of breast cancer and HER2-targeted therapies: a comprehensive review
Medical Oncology (2022)
-
Elevated MMP9 expression in breast cancer is a predictor of shorter patient survival
Breast Cancer Research and Treatment (2020)
-
ErbB1 and ErbB3 co-over expression as a prognostic factor in gastric cancer
Biological Research (2019)