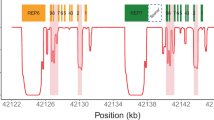
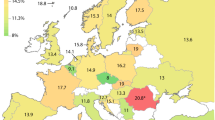
Abstract
The four members of the human CYP3A subfamily play important roles in the clearance of xenobiotics, hormones, and environmental compounds. Many SNPs at the CYP3A locus have been characterized, with several showing large allele frequency differences across populations. In addition to the effects of CYP3A SNPs on drug metabolism, recent studies have highlighted the potential for CYP3A variation in susceptibility to several common phenotypes, including hypertension and cancer. We previously showed that the CYP3A4 and CYP3A5 genes have a strong haplotype structure at varying frequencies across ethnic groups. Here, we extend our re-sequencing survey to the remaining CYP3A genes in the same cluster, CYP3A7 and CYP3A43. Our study identified a large number of SNPs in coding and conserved noncoding sequences, several of which are common. The combined data set allows us to investigate patterns of sequence variation and linkage disequilibrium at the entire CYP3A locus for use in future association studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Halpert JR . Structural basis of selective cytochrome P450 inhibition. Annu Rev Pharmacol Toxicol 1995; 35: 29–53.
Venkatakrishnan K, von Moltke LL, Greenblatt DJ . Effects of the antifungal agents on oxidative drug metabolism: clinical relevance. Clin Pharmacokinet 2000; 38: 111–180.
Mutch E, Daly AK, Leathart JB, Blain PG, Williams FM . Do multiple cytochrome P450 isoforms contribute to parathion metabolism in man? Arch Toxicol 2003; 77: 313–320.
Dorne JL, Walton K, Renwick AG . Human variability in CYP3A4 metabolism and CYP3A4-related uncertainty factors for risk assessment. Food Chem Toxicol 2003; 41: 201–224.
Guengerich FP . Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol 1999; 39: 1–17.
Li AP, Kaminski DL, Rasmussen A . Substrates of human hepatic cytochrome P450 3A4. Toxicology 1995; 104: 1–8.
Evans WE, Relling MV . Pharmacogenomics: translating functional genomics into rational therapeutics. Science 1999; 286: 487–491.
Nebert DW, Russell DW . Clinical importance of the cytochromes P450. Lancet 2002; 360: 1155–1162.
Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ et al. Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 2003; 307: 573–582.
Ohmori S, Nakasa H, Asanome K, Kurose Y, Ishii I, Hosokawa M et al. Differential catalytic properties in metabolism of endogenous and exogenous substrates among CYP3A enzymes expressed in COS-7 cells. Biochim Biophys Acta 1998; 1380: 297–304.
Marill J, Capron CC, Idres N, Chabot GG . Human cytochrome P450s involved in the metabolism of 9-cis- and 13-cis-retinoic acids. Biochem Pharmacol 2002; 63: 933–943.
Leeder JS, Gaedigk R, Marcucci KA, Gaedigk A, Vyhlidal CA, Schindel BP et al. Variability of CYP3A7 expression in human fetal liver. J Pharmacol Exp Ther 2005; 314: 626–635.
Rodriguez-Antona C, Jande M, Rane A, Ingelman-Sundberg M . Identification and phenotype characterization of two CYP3A haplotypes causing different enzymatic capacity in fetal livers. Clin Pharmacol Ther 2005; 77: 259–270.
Westlind A, Malmebo S, Johansson I, Otter C, Andersson TB, Ingelman-Sundberg M et al. Cloning and tissue distribution of a novel human cytochrome p450 of the CYP3A subfamily, CYP3A43. Biochem Biophys Res Commun 2001; 281: 1349–1355.
Stone A, Ratnasinghe LD, Emerson GL, Modali R, Lehman T, Runnells G et al. CYP3A43 Pro(340)Ala polymorphism and prostate cancer risk in African Americans and Caucasians. Cancer Epidemiol Biomarkers Prev 2005; 14: 1257–1261.
Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB . Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst 1998; 90: 1225–1229.
Lamba JK, Lin YS, Schuetz EG, Thummel KE . Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 2002; 54: 1271–1294.
Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 2001; 27: 383–391.
Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 2001; 11: 773–779.
Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A . CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet 2004; 75: 1059–1069.
Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J et al. PipMaker – a web server for aligning two genomic DNA sequences. Genome Res 2000; 10: 577–586.
Ebersberger I, Metzler D, Schwarz C, Paabo S . Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet 2002; 70: 1490–1497.
Cauffiez C, Lo-Guidice JM, Chevalier D, Allorge D, Hamdan R, Lhermitte M et al. First report of a genetic polymorphism of the cytochrome P450 3A43 (CYP3A43) gene: identification of a loss-of-function variant. Hum Mutat 2004; 23: 101.
Wang H, Dick R, Yin H, Licad-Coles E, Kroetz DL, Szklarz G et al. Structure–function relationships of human liver cytochromes P450 3A: aflatoxin B1 metabolism as a probe. Biochemistry 1998; 37: 12536–12545.
Ng PC, Henikoff S . Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863–874.
Ki CS, Lee KA, Lee SY, Kim HJ, Cho SS, Park JH et al. Haplotype structure of the UDP-glucuronosyltransferase 1A1 (UGT1A1) gene and its relationship to serum total bilirubin concentration in a male Korean population. Clin Chem 2003; 49: 2078–2081.
Yamamoto K, Sato H, Fujiyama Y, Doida Y, Bamba T . Contribution of two missense mutations (G71R and Y486D) of the bilirubin UDP glycosyltransferase (UGT1A1) gene to phenotypes of Gilbert's syndrome and Crigler–Najjar syndrome type II. Biochim Biophys Acta 1998; 1406: 267–273.
Springer MS, Murphy WJ, Eizirik E, O'Brien SJ . Placental mammal diversification and the Cretaceous–Tertiary boundary. Proc Natl Acad Sci USA 2003; 100: 1056–1061.
Watterson GA . On the number of segregating sites in genetical models without recombination. Theor Popul Biol 1975; 7: 256–276.
Tajima F . Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989; 123: 585–595.
Akey JM, Zhang G, Zhang K, Jin L, Shriver MD . Interrogating a high-density SNP map for signatures of natural selection. Genome Res 2002; 12: 1805–1814.
Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ . Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 1994; 136: 1329–1340.
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA . Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 2004; 74: 106–120.
Bradlow HL, Hershcopf RJ, Martucci CP, Fishman J . Estradiol 16 alpha-hydroxylation in the mouse correlates with mammary tumor incidence and presence of murine mammary tumor virus: a possible model for the hormonal etiology of breast cancer in humans. Proc Natl Acad Sci USA 1985; 82: 6295–6299.
Kabat GC, Chang CJ, Sparano JA, Sepkovie DW, Hu XP, Khalil A et al. Urinary estrogen metabolites and breast cancer: a case–control study. Cancer Epidemiol Biomarkers Prev 1997; 6: 505–509.
Schneider J, Kinne D, Fracchia A, Pierce V, Anderson KE, Bradlow HL et al. Abnormal oxidative metabolism of estradiol in women with breast cancer. Proc Natl Acad Sci USA 1982; 79: 3047–3051.
Guengerich FP, Shimada T . Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem Res Toxicol 1991; 4: 391–407.
Kadlubar FF . Biochemical individuality and its implications for drug and carcinogen metabolism: recent insights from acetyltransferase and cytochrome P4501A2 phenotyping and genotyping in humans. Drug Metab Rev 1994; 26: 37–46.
Spurdle AB, Goodwin B, Hodgson E, Hopper JL, Chen X, Purdie DM et al. The CYP3A4*1B polymorphism has no functional significance and is not associated with risk of breast or ovarian cancer. Pharmacogenetics 2002; 12: 355–366.
Zheng W, Jin F, Dunning LA, Shu XO, Dai Q, Wen WQ et al. Epidemiological study of urinary 6beta-hydroxycortisol to cortisol ratios and breast cancer risk. Cancer Epidemiol Biomarkers Prev 2001; 10: 237–242.
Henikoff S, Henikoff JG . Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 1992; 89: 10915–10919.
Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA 2003; 100: 5896–5901.
Dai D, Tang J, Rose R, Hodgson E, Bienstock RJ, Mohrenweiser HW et al. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther 2001; 299: 825–831.
Eiselt R, Domanski TL, Zibat A, Mueller R, Presecan-Siedel E, Hustert E et al. Identification and functional characterization of eight CYP3A4 protein variants. Pharmacogenetics 2001; 11: 447–458.
Kajinami K, Brousseau ME, Ordovas JM, Schaefer EJ . CYP3A4 genotypes and plasma lipoprotein levels before and after treatment with atorvastatin in primary hypercholesterolemia. Am J Cardiol 2004; 93: 104–107.
Matsumura K, Saito T, Takahashi Y, Ozeki T, Kiyotani K, Fujieda M et al. Identification of a novel polymorphic enhancer of the human CYP3A4 gene. Mol Pharmacol 2004; 65: 326–334.
Thomas JW, Prasad AB, Summers TJ, Lee-Lin SQ, Maduro VV, Idol JR et al. Parallel construction of orthologous sequence-ready clone contig maps in multiple species. Genome Res 2002; 12: 1277–1285.
Roe BA . Shotgun library construction for DNA sequencing. Methods Mol Biol 2004; 255: 171–187.
Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG et al. Whole-genome patterns of common DNA variation in three human populations. Science 2005; 307: 1072–1079.
Nickerson DA, Tobe VO, Taylor SL . PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 1997; 25: 2745–2751.
Hill WG . Estimation of linkage disequilibrium in randomly mating populations. Heredity 1974; 33: 229–239.
Stephens M, Smith NJ, Donnelly P . A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989.
Weir BS . Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer Associates Inc.: Sunderland, MA, 1996.
Acknowledgements
We thank members of the Pharmacogenetics of Anticancer Agents Research (PAAR) group for helpful discussions throughout the project, particularly E Schuetz. We thank D Nickerson and J Akey for providing polymorphism and divergence data for the Seattle SNPs genes. EET was partially supported by training grant GM07197. Portions of this work were supported by NIH Grant 5 U54 HG02152 to BAR. This Pharmacogenetics of Anticancer Agents Research (PAAR) Group study was supported by the NIH/NIGMS Grant U01GM61393, www.pharmacogenetics.org. Data will be deposited into PharmGKB (supported by NIH/NIGMS Pharmacogenetics Research Network and Database Grant U01GM61374, http://pharmgkb.org/).
Author information
Authors and Affiliations
Corresponding author
Additional information
Duality of Interest
None declared.
Electronic Database Information
Cluster-Buster, http://zlab.bu.edu/cluster-buster/
Coriell Cell Repositories, http://locus.umdnj.edu/ccr/
Di Rienzo Lab Web site, http://genapps.uchicago.edu/labweb/pubs.html (for primer sequences, population sample informa tion, and data)
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CYP3A locus (accession number NG_000004) and individual gene accession numbers for CYP3A43 (AC011904), CYP3A4 and CYP3A7 (AC069294), and CYP3A5 (AC005020))
MultiPipMaker, http://bio.cse.psu.edu/pipmaker
PharmGKB, http://www.pharmgkb.org/ (for primer sequences and population samples used in re-sequencing study (individual gene accession numbers for CYP3A43 (PS205160), CYP3A4 (PS203894), CYP3A7 (PS205159), and CYP3A5 (PS203895))
Roe lab web site, http://www.genome.ou.edu/proto.html (for the most recent versions of the protocols used for BAC isolation, random shotgun cloning and sequencing, and sequence finishing)
SLIDER, http://genapps.uchicago.edu/slider/index.html (for computing summary statistics of population genetic data)
University of Washington-Fred Hutchinson Cancer Research Center, http://pga.gs.washington.edu/education.html (for Seattle SNPs, the National Heart Lung and Blood Institute's Program for Genomic Applications)
SIFT (Sorting Intolerant From Tolerant), http://blocks.fhcrc.org/sift/SIFT.html
Multi-Lagan, http://lagan.stanford.edu/cgi-bin/MLaganInput.cgi (for performing sequence alignments)
MATCH 1.0, www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi (for prediction of transcription factor binding sites)
MAXDIP, http://genapps.uchicago.edu/maxdip/index.html (to generate composite likelihood estimates of recombination rate based on LD data)
Supplementary Information accompanies the paper on The Pharmacogenomics Journal website (http://www.nature.com/tpj).
Supplementary information
Rights and permissions
About this article
Cite this article
Thompson, E., Kuttab-Boulos, H., Yang, L. et al. Sequence diversity and haplotype structure at the human CYP3A cluster. Pharmacogenomics J 6, 105–114 (2006). https://doi.org/10.1038/sj.tpj.6500347
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.tpj.6500347
Keywords
This article is cited by
-
Whole exome sequencing for the identification of CYP3A7 variants associated with tacrolimus concentrations in kidney transplant patients
Scientific Reports (2018)
-
A pharmacogenomic study on the pharmacokinetics of tacrolimus in healthy subjects using the DMETTM Plus platform
The Pharmacogenomics Journal (2017)
-
Exploring the relationship between lifestyles, diets and genetic adaptations in humans
BMC Genetics (2015)
-
Metabolism of aflatoxins: key enzymes and interindividual as well as interspecies differences
Archives of Toxicology (2014)
-
Molecular diversity and population structure at the Cytochrome P450 3A5 gene in Africa
BMC Genetics (2013)