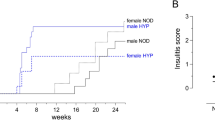
Abstract
While sharing the H2g7 MHC and many other important Type I diabetes susceptibility (Idd) genes with NOD mice, the NOR strain remains disease free due to resistance alleles within the ∼12% portion of their genome that is of C57BLKS/J origin. Previous F2 segregation analyses indicated multiple genes within the ‘Idd13’ locus on Chromosome 2 provide the primary component of NOR diabetes resistance. However, it was clear other genes also contribute to NOR diabetes resistance, but were difficult to detect in the original segregation analyses because they were relatively weak compared to the strong Idd13 protection component. To identify these further genetic components of diabetes resistance, we performed a new F2 segregation analyses in which NOD mice were outcrossed to a ‘genome-conditioned’ NOR stock in which a large component of Idd13-mediated resistance was replaced with NOD alleles. These F2 segregation studies combined with subsequent congenic analyses confirmed the presence of additional NOR resistance genes on Chr. 1 and Chr. 4, and also potentially on Chr. 11. These findings emphasize the value for diabetes gene discovery of stratifying not only MHC loci conferring the highest relative risk but also as many as possible of the non-MHC loci presumed to contribute significantly.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Serreze DV, Leiter EH . Genes and pathways underlying autoimmune diabetes in NOD mice. In: von Herrath M (ed). Molecular Pathology of Insulin-Dependent Diabetes Mellitus. Karger: New York, 2001, pp 31–67.
Prochazka M, Serreze DV, Frankel WN, Leiter EH . NOR/Lt; MHC-matched diabetes-resistant control strain for NOD mice. Diabetes 1992; 41: 98–106.
Serreze DV, Prochazka M, Reifsnyder PC, Bridgett MM, Leiter EH . Use of recombinant congenic and congenic strains of NOD mice to identify a new insulin dependent diabetes resistance gene. J Exp Med 1994; 180: 1553–1558.
Naggert JK, Mu M-L, Frankel WF, Paigen B . Genomic analysis of the C57BL/Ks mouse strain. Mammal Genome 1995; 6: 131–133.
McAleer MA, Reifsnyder P, Palmer SM et al. Crosses of NOD mice with the related NON strain: a polygenic model for type I diabetes. Diabetes 1995; 44: 1186–1195.
Wicker LS, Todd JA, Peterson LB . Genetic control of autoimmune diabetes in the NOD mouse. Ann Rev Immunol 1995; 13: 179–200.
Serreze DV, Bridgett MB, Chapman HD, Chen E, Richard SB, Leiter EH . Subcongenic analysis of the Idd13 locus in NOD/Lt mice: evidence for several susceptibility genes including a possible diabetogenic role for β2-microglobulin. J Immunol 1998; 160: 1472–1478.
Hamilton-Williams EE, Serreze DV, Charlton B et al. Transgenic rescue implicates β2-microglobulin as a diabetes susceptibility gene in NOD mice. Proc Natl Acad Sci USA 2001; 98: 11533–11538.
Fox CJ, Danska JS . Independent genetic regulation of T-cell and antigen-presenting cell participation in autoimmune islet inflammation. Diabetes 1998; 47: 331–338.
Fox CJ, Paterson AD, Mortin-Toth SM, Danska JS . Two genetic loci regulate T cell-dependent islet inflammation and drive autoimmune diabetes pathogenesis. Am J Hum Genet 2000; 67: 67–81.
Morahan G, McClive P, Huang D, Little P, Baxter A . Genetic and physiological association of diabetes susceptibility with raised Na+/H+ exchange activity. Proc Natl Acad Sci USA 1994; 91: 5898–5902.
Brodnicki TC, McClive P, Couper S, Morahan G . Localization of Idd11 using NOD congenic mouse strains: elimination of Slc9a1 as a candidate gene. Immunogenetics 2000; 51: 37–41.
Lyons PA, Hancock WW, Denny P et al. The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity 2000; 13: 107–115.
Petkov PM, Ding Y, Cassell MA et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res 2004; 14: 1806–1811.
Ueda H, Howson JM, Esposito L et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003; 423: 506–511.
Todd JA, Aitman TJ, Cornall RJ et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature 1991; 351: 542–547.
Ghosh S, Palmer SM, Rodrigues NR et al. Polygenic control of autoimmune diabetes in nonobese diabetic mice. Nat Genet 1993; 4: 404–409.
Wicker LS, Todd JA, Prins J-B, Podolin PL, Renjilian RJ, Peterson LB . Resistance alleles in two non-MHC-linked insulin dependent diabetes loci on chromosome 3, Idd3 and Idd10, protect NOD mice from diabetes. J Exp Med 1994; 180: 1705–1713.
Grattan M, Mi QS, Meagher C, Delovitch TL . Congenic mapping of the diabetogenic locus Idd4 to a 5.2-cM region of chromosome 11 in NOD mice: identification of two potential candidate subloci. Diabetes 2002; 51: 215–223.
Simpson PB, Mistry MS, Maki RA et al. Cutting edge: diabetes-associated quantitative trait locus, Idd4, is responsible for the IL-12p40 overexpression defect in nonobese diabetic (NOD) mice. J Immunol 2003; 171: 3333–3337.
McDuffie M . Derivation of diabetes-resistant congenic lines from the nonobese diabetic mouse. Clin Immunol 2000; 96: 119–130.
Hill NJ, Lyons PA, Armitage N, Todd JA, Wicker LS, Peterson LB . The NOD Idd5 locus controls insulitis and diabetes and overlaps the orthologous CTLA4/IDDM12 and NRAMP1 loci in humans. Diabetes 2000; 49: 1744–1747.
Verdaguer J, Amrani A, Anderson B, Schmidt D, Santamaria P . Two mechanisms for the non-MHC-linked resistance to spontaneous autoimmunity. J Immunol 1999; 162: 4614–4626.
Stratmann T, Martin-Orozco N, Mallet-Designe V et al. Susceptible MHC alleles, not background genes, select an autoimmune T cell reactivity. J Clin Invest 2003; 112: 902–914.
Baxter AG, Smyth MJ . The role of NK cells in autoimmune disease. Autoimmunity 2002; 35: 1–14.
Matsuki N, Stanic AK, Embers ME, Van Kaer L, Morel L, Joyce S . Genetic dissection of V alpha 14J alpha 18 natural T cell number and function in autoimmune-prone mice. J Immunol 2003; 170: 5429–5437.
Esteban LM, Tsoutsman T, Jordan MA et al. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol 2003; 171: 2873–2878.
Sen P, Bhattacharyya S, Wallet M et al. NF-kappa B hyperactivation has differential effects on the APC function of nonobese diabetic mouse macrophages. J Immunol 2003; 170: 1770–1780.
Wicker LS, Chamberlain G, Hunter K et al. Fine mapping, gene content, comparative sequencing, and expression analyses support Ctla4 and Nramp1 as candidates for Idd51 and Idd52 in the nonobese diabetic mouse. J Immunol 2004; 173: 164–173.
Lamhamedi-Cherradi SE, Boulard O, Gonzalez C et al. Further mapping of the Idd51 locus for autoimmune diabetes in NOD mice. Diabetes 2001; 50: 2874–2878.
Leiter E . The NOD mouse: a model for insulin dependent diabetes mellitus. In: Coligan JE, Kruisbeek AM, Margulies DM, Shevach EM, Strober W (eds). Current Protocols in Immunology. John Wiley & Sons Inc.: New York, 1997, pp 15.19.11–15.19.23.
Sen S, Churchill GA . A statistical framework for quantitative trait mapping. Genetics 2001; 159: 371–387.
Lander ES, Bottstein D . Mapping complex genetic traits in humans: new methods using a complete RFLP linkage map. Cold Spring Harb Symp Quant Biol 1986; LI: 49–62.
Churchill GA, Doerge RW . Empirical threshold value for quantitative trait mapping. Genetics 1994; 138: 963–971.
Sugiyama F, Churchill GA, Higgins DC et al. Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics 2001; 71: 70–77.
Acknowledgements
We gratefully acknowledge the assistance provided by Pamela Stanley, Bruce Regimbal and Steve Langley for animal breeding, and by Gunjan Wagner for assistance with SNP genotyping. This work was supported by NIH Grants DK27722 and DK36175. Institutional shared services were supported by National Cancer Institute Center Support Grant CA-34196.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reifsnyder, P., Li, R., Silveira, P. et al. Conditioning the genome identifies additional diabetes resistance loci in Type I diabetes resistant NOR/Lt mice. Genes Immun 6, 528–538 (2005). https://doi.org/10.1038/sj.gene.6364241
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gene.6364241
Keywords
This article is cited by
-
Diabetic modifier QTLs in F2 intercrosses carrying homozygous transgene of TGF-β
Mammalian Genome (2008)
-
Subcongenic analysis of genetic basis for impaired development of invariant NKT cells in NOD mice
Immunogenetics (2007)