Abstract
Crigler–Najjar syndrome type I (CN-I) is a rare and severe metabolic disorder. A recurrent mutation – c.1070A>G in exon 3 – was identified in the Tunisian population, suggesting a founder effect. In 2004, the detection of this mutation in two Kuwaiti Bedouin families has called the Tunisian founder effect in question again. To determine the origin of this mutation, 21 Tunisian and 2 Kuwaiti Bedouin CN-I patients were screened using nine genetic markers. Haplotype analysis confirmed the founder effect hypothesis and dated the appearance of this mutation some 32 generations ago in the Tunisian population. Using the same genetic analysis, the ancestor haplotype was identified in these two families. This result genetically confirms the blending of the Bedouin nomads within today's Tunisian population. After population migration from east to west, this mutation was introduced into the Tunisian population, and then perpetuated, probably because of marriages in isolated communities.
Similar content being viewed by others
Introduction
Crigler–Najjar syndrome type I (CN-I, MIM no. 218800) is a rare and severe autosomal disorder (less than 1/106 live births). CN-I is caused by deficiency of the liver enzyme responsible for bilirubin elimination, the uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1; EC 2.4.1.17). Biologically, the disease manifests itself with severe and persistent unconjugated hyperbilirubinemia. Clinically, affected newborns are at high risk for bilirubin-induced brain damage (kernicterus). The UGT1A locus complex on chromosome 2 (located on 2q37.1) encodes nine functional UDP-glucuronosyltransferases type 1A.1, 2 The locus is organized with 13 different exons 1 (responsible for the specificity of the enzyme activity) and 4 common exons. Recently, a new regulation system of the UGT1A1 protein activity was identified with an alternative exon 5.3 As for the UGT1A1 gene, about 70 different mutations have been reported since 1992, in the five exons or in splicing sites.4, 5, 6 In our laboratory, the recurrent mutation c.1070A>G responsible for the modification in the peptide sequence p.Gln357Arg has been only observed in patients from Tunisia, suggesting a Tunisian founder effect.7 Recently, this mutation has also been reported in two Kuwaiti Bedouin families, suggesting a wider Arabian founder effect.8
The recurrence of a particular mutation may result from three different conditions. First, the mutation occurs in a region of high risk for mutation (‘hot spot’), such as CpG dinucleotides.9 Second, an external event (such as an infectious disease) has exerted a pressure on the population and selected individuals carrying the morbid allele (eg, the mutation responsible for sickle cell anemia was preserved in the African population by conferring on heterozygote patients, a resistance to malaria).10 Finally, the mutation appeared in one individual and diffused through generations in isolated communities because of consanguineous marriages. In this situation, the chromosomal fragment bearing the mutation is transmitted from a common ancestor to many persons in successive generations. When a founder effect is suspected, several tandem repeats or microsatellites must be analyzed in a restricted genomic region to confirm the single origin (common ancestor) of a recurrent mutation in a population. During meiosis, chromosomal recombination can randomly occur, so that the more recent the mutation is, the more conserved the genetic region around the mutation will be. In these cases, microsatellites around the mutation will disclose the same or at least a similar haplotype in all affected patients.
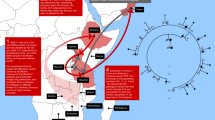
In this study, genetic markers on both centromeric and telomeric sides of the c.1070A>G mutation in the UGT1A1 gene were analyzed in 21 Tunisian and 2 Kuwaiti CN-I patients. We have determined that this mutation probably appeared in a Bedouin nomad group and then diffused in Tunisia around eight centuries ago after population migrations from Middle East to Maghreb.
Patients and methods
To determine the origin of the c.1070A>G mutation in the UGT1A1 gene, 28 healthy unrelated Tunisians (group 1), 21 CN-I Tunisian patients (group 2) and 2 CN-I Kuwaiti patients (group 3) were included. Seven microsatellite markers (centromere-D2S2344, D2S331, D2S1279, D2S2348, D2S2234, D2S2205 and D2S336-telomere) were selected from the Ensembl Genome Project Database. Haplotypes were constructed by amplification of microsatellites with fluorescent primers 5′-labeled in a multiplex PCR except for D2S336 and D2S2205, each amplified in a single PCR. The samples were run on an ABI PRISM 3130 DNA analyzer (Applied Biosystems). The results were analyzed by using Genescan 3.1 analysis software to determine the PCR fragment sizes of PCR products, and the allele sizes were carried out automatically with Genotyper 2.5 analysis software. Single PCRs were performed using the same program for D2S336 and D2S2205. Haplotypes were classified as 1 for the shortest to 20 for the longest.
To complete this panel, two other markers were added: the UGT1A1 TATA box polymorphism and a new polymorphic genetic marker ∼77 000 bp upstream from UGT1A1. The polymorphism A(TA)6TAA or A(TA)7TAA in the promoter of the UGT1A1 gene was determined by PCR followed by electrophoresis on polyacrylamide gel as described previously.11 The second genetic marker – called MARK01 – was a CA-repeat sequence whose variability had previously been assayed on 10 French healthy unrelated volunteers. Its physical location was calculated to be 77 440 pb upstream from the mutation c.1070A>G. Primers to amplify this specific region were designed using Primer3 (Primer3 web site: http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The primer sequences were as follows: forward primer 5′-GGAGCTACTCTTTAGGGATCG-3′; reverse primer 5′-TGTGAGCTTTGACTGTACTAAG-3′. After PCR amplification, fragments were separated by electrophoresis onto a 10% polyacrylamide gel and then were visualized after ethidium bromide staining. As for the other microsatellites studied, haplotypes were classified according to their electrophoretic profile as 1 for the shortest to 4 for the longest.
To determine linkage disequilibrium (LD) score, the appropriate physical-to-genetic distance conversion relation was determined by linear regression analysis (SPSSv8.0 program, SPSS) of genetic versus physical map position.12 Megabase (UniSTS, NCBI) and centimorgan (Marshfield Comprehensive Human Genetic Map) information for 14 markers positioned in a 10-Mb genomic region around UGT1A1 gene was used (Table 1). Applying this relation, genetic distances were calculated in relation to the mutation c.1070A>G for the nine studied genetic markers (Table 2). Kosambi's function was applied to convert genetic distance (cM) into recombination fraction (θ): θ=0.5(e(cM/25)−1)/(e(cM/25)+1) (Table 2).13 Statistical comparison of allele frequencies between disease and normal chromosomes was based on Mantel–Haenszel common odds ratio estimate with alleles classified into two groups: one for the associated allele and all others combined into a single group. The LD was calculated applying the formula δ=(p11p22−p12p21)/(p+1p22), where p11 is the frequency of the associated allele on disease chromosomes, p22 the frequency of the normal alleles on normal chromosomes, p12 the frequency of the associated allele on normal chromosomes, p21 the frequency of the normal alleles on disease chromosomes and p+1 the frequency of disease chromosomes.13
The number of generations (g) since the appearance of the mutation was calculated with the formula g=ln(δ)/ln(θ−1), where δ is the LD and θ the recombination fraction.14
Results
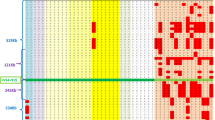
Allele frequencies for the nine genetic markers group are presented in Tables 3a, b (for group 1 and group 2, respectively).
The polymorphism observed in the CN-I Tunisian population was strongly restricted in comparison with the healthy Tunisian population (Table 2). For example, for D2S2348, only 4 different alleles out of 20 observed in the healthy Tunisian population were identified in the CN-I Tunisian population. In the healthy Tunisian population, frequency of heterozygotes for the A(TA)7TAA allele was 0.393 and frequency of homozygotes was 0.178. For all Tunisian CN-I patients, the c.1070A>G mutation was associated with the homozygous A(TA)7TAA mutant allele in the promoter. For MARK01, the allele 4 frequency was 0.952 in the Tunisian CN-I population and only 0.357 in the healthy Tunisian population. The ancestor haplotype (D2S2344: 5, D2S331: 9, D2S1279: 9, D2S2348: 5, D2S2234: 5, D2S2205: 1, MARK01: 4, TATA box: A(TA)7TAA and D2S336: 4) was observed in three Tunisian CN-I patients and was not observed in the healthy Tunisian population.
LD calculated with the Delvin and Risch formula decreased from 1.000 to 0.4524 with increasing distance of markers from the mutation, except for D2S2234 and D2S2348. The age of the mutation was estimated to be between 21 and 60 generations (mean 32 generations).
In the two Kuwaiti patients, the c.1070A>G mutation was also associated with the homozygous A(TA)7TAA mutant allele in the promoter. Their haplotype was identical to the ancestor one.
Discussion
CN-I is a rare genetic disease whose frequency is estimated to be 1/106 births and it affects boys and girls in the same proportion. Founder effects have already been suspected for CN-I in isolated communities such as in France, Portugal or Sardinia.15, 16
In our laboratory, clinical and/or biochemical CN-I was genetically confirmed for 56 patients. Twenty-five of these patients originated from different part of Tunisia (Tunis, Sfax and Sousse) and 21 were homozygous for the c.1070A>G mutation in exon 3 associated with the A(TA)7TAA/A(TA)7TAA polymorphism in the TATA box (three of the other patients were homozygous for the deletion c.396_401delCAACAA associated with the A(TA)7TAA/A(TA)7TAA polymorphism and the fourth was homozygous for a large deletion including the promoter and the exon 1). The implication of this mutation in the CN-I phenotype has never been determined by in vitro expression studies, but convincing arguments support this relation, particularly the frequency of this mutation in the CN-I Tunisian population. However, the c.1070A>G mutation has always been found associated with the A(TA)7TAA/A(TA)7TAA polymorphism, so that it is, to date, impossible to determine whether this mutation totally abolishes UGT1A1 activity by itself or whether its association with the A(TA)7TAA/A(TA)7TAA genotype in the TATA box is necessary. In the Tunisian population, the Gilbert allele A(TA)7TAA frequency was found to be 0.393 and 0.178 for heterozygotes and homozygotes, respectively. These data were comparable to those observed in France (0.385 and 0.17, respectively) or Greece (0.328 and 0.186, respectively).17, 18
CN-I or II cases have been described in several regions of the Middle East and the Maghreb such as Morocco, Algeria, Tunisia, Saudi Arabia, Lebanon or Kuwait.7, 8, 15, 19, 20 The high prevalence of this disease, particularly in Saudi Arabia or Tunisia can be explained by a high rate of consanguineous marriages in restricted communities.19 In the Tunisian population, the large prevalence of the c.1070A>G mutation in exon 3 had suggested an ancestral common origin.7 The absence of particular susceptibility of the exon 3 to this mutation (only observed in Tunisia and in two Kuwaiti Bedouins families) reinforces the founder effect hypothesis.7, 8 On the other hand, the question of a protective effect of Crigler–Najjar causing alleles at the heterozygote state against oxidative damages such as heart coronary diseases is not clear. Vitek et al21 pointed that patients with mild increase of serum bilirubin could be protected against heart disease. But on the other hand, Bosma et al22 and Gajdos et al23 in two different studies did not find any correlation between mutations in UGT1A1 gene promoter and protection against heart disease. Moreover an eventual protective effect would be observed after reproductive age. These observations could not explain the high prevalence of Crigler–Najjar syndrome in the Tunisian population.
In this study, genetic analysis allowed us to confirm for the first time the founder effect hypothesis in the Tunisian population. The c.1070A>G mutation appeared some 32 generations ago (nearly eight centuries ago), and its diffusion was strongly limited, probably thanks to consanguineous marriages. The systematic presence of the A(TA)7TAA/A(TA)7TAA genotype in the promoter in c.1070A>G mutation carriers suggests that this mutation appeared after the promoter polymorphism, reinforcing the founder effect hypothesis. Genetically, the mutation c.1070A>G and the TATA box are very close (7702 bp or 0.021 cM). In a population of 21 patients, the number of generations necessary to observe one meiotic recombination between these two points is estimated to be 191, spanning about 4000 years.
Unfortunately, the Kuwaiti population studied was too restricted to determine LD, but genetic marker analysis allowed us to identify the ancestor haplotype in the two patients.
Referring to the Bedouin population history, the mutation probably appeared in the Bedouin community and was introduced in the Tunisian population nearly eight centuries ago after human migrations from east to west. Indeed, Bedouin nomads spread out from the Arabian Peninsula into all countries between the Arabic Gulf and the Atlantic. Bedouins arrived in Tunisia very early during the first Arab-Muslim invasions and married the native population, leading to today's population.
References
Ritter JK, Chen F, Sheen YY et al: A novel complex locus UGT1 encodes human bilirubin, phenol, and other UDP-glucuronosyltransferase isoenzymes with identical carboxyl termini. J Biol Chem 1992; 267: 3257–3261.
Gong QH, Cho JW, Huang T et al: Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics 2001; 11: 357–368.
Lévesque E, Girard H, Journault K, Lépine J, Guillemette C : Regulation of the UGT1A1 bilirubin-conjugating pathway: role of a new splicing event at the UGT1A locus. Hepatology 2007; 45: 128–138.
Bosma PJ, Chowdhury NR, Goldhoorn BG et al: Sequence of exons and the flanking regions of human bilirubin-UDP-glucuronosyltransferase gene complex and identification of a genetic mutation in a patient with Crigler–Najjar syndrome, type I. Hepatology 1992; 15: 941–947.
Ritter JK, Yeatman MT, Ferreira P, Owens IS : Identification of a genetic alteration in the code for bilirubin UDP-glucuronosyltransferase in the UGT1 gene complex of a Crigler–Najjar type I patient. J Clin Invest 1992b; 90: 150–155.
Kadakol A, Ghosh SS, Sappal BS, Sharma G, Chowdhury JR, Chowdhury NR : Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler–Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum Mutat 2000; 16: 297–306.
Francoual J, Rivierre A, Mokrani C et al: Crigler–Najjar syndrome type I in Tunisia may be associated with a founder effect related to the Q357R mutation within the UGT1 gene. Hum Mutat 2002; 19: 570–571.
Koshy A, Bosma PJ, Oude-Elferink RP : Crigler–Najjar syndrome type 1 associated with combined 1070A>G, Q357R and (TA)7 mutation in Kuwaiti Bedouin families indicate a founder effect in Arabs. J Clin Gastroenterol 2004; 38: 465–467.
Cooper DN, Youssoufian H : The CpG dinucleotide and human genetic disease. Hum Genet 1998; 78: 151–155.
Allison AC : The distribution of the sickle-cell trait in East Africa and elsewhere, and its apparent relationship to the incidence of subtertian malaria. Trans R Soc Trop Med Hyg 1954; 48: 312–318.
Le Bihan-Levaufre B, Francoual J, Labrune P, Chalas J, Capel L, Lindenbaum A : [Refinement and role of the diagnosis of Gilbert disease with molecular biology]. Ann Biol Clin (Paris) 2001b; 59: 61–66.
Colombo R, Bignamini AA, Carobene A et al: Age and origin of the FCMD 3′-untranslated-region retrotransposal insertion mutation causing Fukuyama-type congenital muscular dystrophy in the Japanese population. Hum Genet 2000; 107: 559–567.
Delvin B, Risch N : A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 1995; 29: 311–322.
Risch N, de Leon D, Ozelius L et al: Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet 1995; 9: 152–159.
Labrune P, Myara A, Hadchouel M et al: Genetic heterogeneity of Crigler–Najjar syndrome type I: a study of 14 cases. Hum Genet 1994; 94: 693–697.
Rosatelli MC, Meloni A, Faa V et al: Molecular analysis of patients of Sardinian descent with Crigler–Najjar syndrome type I. J Med Genet 1997; 34: 122–125.
Le Bihan-Levaufre B, Francoual J, Chalas J et al: [Genetic incidence of Gilbert's syndrome in France]. Gastroenterol Clin Biol 2001a; 25: 557–558.
Kavazarakis E, Tsezou A, Tzetis M et al: Gilbert syndrome: analysis of the promoter region of the uridine diphosphate-glucuronosyltransferase 1 gene in the Greek population. Eur J Pediatr 2000; 159: 873–874.
Al Shurafa H, Wali S, Chehab MS et al: Living-related liver transplantation for Crigler–Najjar syndrome in Saudi Arabia. Clin Transplant 2002; 16: 222–226.
Petit FM, Gajdos V, Francoual J et al: Allelic heterogeneity of Crigler–Najjar type I syndrome: a study of 24 cases. Clin Genet 2004; 66: 571–572.
Vítek L, Novotný L, Sperl M, Holaj R, Spácil J : The inverse association of elevated serum bilirubin levels with subclinical carotid atherosclerosis. Cerebrovasc Dis 2006; 21: 408–414.
Bosma PJ, van der Meer IM, Bakker CT, Hofman A, Paul-Abrahamse M, Witteman JC : UGT1A1*28 allele and coronary heart disease: the Rotterdam Study. Clin Chem 2003; 49: 1180–1181.
Gajdos V, Petit FM, Perret C et al: Further evidence that the UGT1A1*28 allele is not associated with coronary heart disease: the ECTIM Study. Clin Chem 2006; 52: 2313–2314.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Petit, F., Bézieau, S., Gajdos, V. et al. The Tunisian population history through the Crigler–Najjar type I syndrome. Eur J Hum Genet 16, 848–853 (2008). https://doi.org/10.1038/sj.ejhg.5201989
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201989
Keywords
This article is cited by
-
Founder mutations in Tunisia: implications for diagnosis in North Africa and Middle East
Orphanet Journal of Rare Diseases (2012)