Abstract
We have investigated a consanguineous Iranian family with eight patients who suffer from mental retardation, disturbed equilibrium, walking disability, strabismus and short stature. By autozygosity mapping we identified one region with a significant LOD score on chromosome 9(p24.2–24.3). The interval contains the VLDLR gene, which codes for the very low-density lipoprotein receptor. This protein is part of the reelin signalling pathway, which is involved in neuroblast migration in the cerebral cortex and cerebellum. A homozygous deletion encompassing VLDLR has previously been found to cause a syndrome of cerebellar ataxia and mental retardation associated with cerebellar hypoplasia in the Hutterite population known as dysequilibrium syndrome (DES). The reported deletion however, contains an additional brain expressed gene of unknown function, whose involvement in the aetiology of the phenotype could so far not be excluded. We screened the coding region of VLDLR for mutations in our patients and found a homozygous c.1342C>T nucleotide substitution, which leads to a premature stop codon in exon 10. This is the first report of a mutation in patients with DES that affects VLDLR exclusively, confirming the central role of the very low-density lipoprotein receptor in the aetiology of this condition.
Similar content being viewed by others
Introduction
Dysequilibrium syndrome (DES, OMIM 224050) is a rare autosomal recessive non-progressive cerebellar disorder characterized by ataxia, mental retardation, cerebellar hypoplasia and—in some patients—strabismus, seizures and short stature.1 Previously, a homozygous ∼199 kb deletion encompassing the entire coding region of the VLDLR gene was detected in several DES patients from the Hutterite population in North America.2 As this deletion also contains an additional brain expressed gene (LOC401491), the authors could not entirely rule out the possibility that this gene might also be contributing to the phenotype. What is more, the size of the deletion did not allow excluding the causal involvement of intergenic regulatory elements. We now describe the detection of a homozygous stop mutation in the VLDLR gene in a large consanguineous Iranian family with DES, providing the first evidence that VLDLR deficiency is exclusively responsible for the DES phenotype.
Clinical report
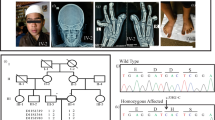
The family was recruited within the framework of a continuous collaborative project that aims at the elucidation of the genetic background of mental retardation in Iran.3 The pedigree is shown in Figure 1. Eight family members (IV:4; IV:5; IV:6; V:1; V:2; V:3; V:4; V:5) suffered from moderate to severe mental retardation. They had either no speech at all or spoke only few words. Motor development was also retarded, they were able to sit independently between the ages of 12 and 24 months, but no patient could walk independently. All patients had strabismus and a body height below the 25th centile; five patients had a body height at or below the 3rd centile. Head circumferences were normal, and neither dysmorphic facial features nor other malformations were observed. Seizures have not been reported for any of the patients. Unfortunately, no images of the brain by MRI or CT scan could be obtained, and putative cerebellar hypoplasia can only be inferred from the neurological defects. The clinical features are summarized in Table 1.
Materials and methods
Sampling procedures and autozygosity mapping procedures were carried out as described previously.3, 4 All investigations were carried out with the written consent of the parents.
For VLDLR mutation screening, primer sequences (available upon request) were generated for all coding and adjacent splice site regions using the ExonPrimer software (http://ihg.gsf.de/ihg/ExonPrimer.html). PCR amplification was carried out using a standard touchdown PCR protocol. After successful amplification PCR products were sequenced on an ABI sequencer (Applied Biosystems, Foster City, CA, USA). For sequence analysis we used the CodonCode Aligner software (CodonCode corporation, Dedham, MA, USA).
Results and discussion
Autozygosity mapping in this family led to the identification of a single linkage interval (one LOD down method), on chromosome 9. This analysis was performed under the assumption of autosomal recessive inheritance with complete penetrance. The region on chromosome 9p24.2–24.3, which comprised approximately 3.7 Mb between the flanking markers rs1532309 and rs4131424 contains the VLDLR gene. As VLDLR has previously been implicated in the aetiology of DES2 we selected it for mutation screening and found a homozygous c.1342C>T nucleotide substitution (Figure 2), which introduces a premature stop codon in exon 10 (R448X). This change cosegregates with the disease and was excluded in 100 healthy Iranian controls by PCR and sequencing.
Nonsense mutation in exon 10 of VLDLR. The gene structure of VLDLR is shown, with numbered boxes representing the exons. Red shading indicates coding regions, blue shading marks untranslated regions. Chromatograms represent sequence fragments of the wildtype (Wt) and mutant (Mut) allele. The affected codon is indicated.
VLDLR consists of 19 exons and encodes a 873 amino acid receptor protein of the low-density lipoprotein receptor superfamily.5 Two isoforms of the protein are known, the full-length version (type I) and a version lacking an O-linked sugar region (type II).6 Type I is mainly distributed in heart and skeletal muscles with active fatty acid metabolism, whereas VLDLR type II is predominant in non-muscular tissue, including kidney, spleen, adrenal gland, lung, brain, testis, uterus and ovary, but not the liver.7, 8 Like the deletion carriers in the Hutterite family,2 our patients can be considered to lack a functional VLDLR transcript, since the stop mutation we report affects both isoforms. The differences in tissue distribution together with divergent ligand specificity between the two isoforms suggest that they play distinct roles in various tissues and cells. As the DES phenotype comprises mostly central nervous system features, it has to be assumed that differences in compensation of VLDLR deficiency are responsible for the functional integrity of the other tissues. In the brain, VLDLR is part of the reelin signalling pathway, which contributes to the correct regulation of neuronal migration for example, by guiding neuroblast migration in the developing cerebral cortex and cerebellum.9, 10, 11 As discussed previously by Boycott et al,2 a possible explanation for the DES phenotype might therefore be that neurons in the cortex fail to distribute normally after reaching their assigned layer, as observed in Vldlr-deficient mice.12
In summary, we report the first DES family outside the Hutterite population and our results show a deleterious mutation that exclusively affects VLDLR in patients with clinical features that are indistinguishable from previously reported cases with a complete lack of VLDLR. Whereas, the size of the earlier reported deletion could not exclude an additional contribution to the genotype from neighbouring genes, our finding clearly demonstrates that VLDLR deficiency alone is sufficient to cause the human DES phenotype.
References
Glass HC, Boycott KM, Adams C et al: Autosomal recessive cerebellar hypoplasia in the Hutterite population. Dev Med Child Neurol 2005; 47: 691–695.
Boycott KM, Flavelle S, Bureau A et al: Homozygous deletion of the very low-density lipoprotein receptor gene causes autosomal recessive cerebellar hypoplasia with cerebral gyral simplification. Am J Hum Genet 2005; 77: 477–483.
Najmabadi H, Motazacker MM, Garshasbi M et al: Homozygosity mapping in consanguineous families reveals extreme heterogeneity of non-syndromic autosomal recessive mental retardation and identifies 8 novel gene loci. Hum Genet 2006; 121: 43–48.
Garshasbi M, Motazacker MM, Kahrizi K et al: SNP array-based homozygosity mapping reveals MCPH1 deletion in family with autosomal recessive mental retardation and mild microcephaly. Hum Genet 2006; 118: 708–715.
Strickland DK, Kounnas MZ, Argraves WS : LDL receptor-related protein: a multiligand receptor for lipoprotein and proteinase catabolism. FASEB J 1995; 9: 890–898.
Sakai J, Hoshino A, Takahashi S et al: Structure, chromosome location, and expression of the human very low-density lipoprotein receptor gene. J Biol Chem 1994; 269: 2173–2182.
Takahashi S, Suzuki J, Kohno M et al: Enhancement of the binding of triglyceride-rich lipoproteins to the very low-density lipoprotein receptor by apolipoprotein E and lipoprotein lipase. J Biol Chem 1995; 270: 15747–15754.
Webb JC, Patel DD, Jones MD, Knight BL, Soutar AK : Characterization and tissue-specific expression of the human ‘very low density lipoprotein (VLDL) receptor' mRNA. Hum Mol Genet 1994; 3: 531–537.
D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T : A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 1995; 374: 719–723.
Rice DS, Curran T : Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci 2001; 24: 1005–1039.
Tissir F, Goffinet AM : Reelin and brain development. Nat Rev Neurosci 2003; 4: 496–505.
Trommsdorff M, Gotthardt M, Hiesberger T et al: Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 1999; 97: 689–701.
Acknowledgements
We are grateful to the patients and their family for their cooperation. Furthermore, we acknowledge the contributions of Sahar Esmaeeli Nieh, Seyedeh Sedigheh Abedini, Sanaz Arzhangi as well as Sussan Banihashemi and thank Bettina Lipkowitz for technical assistance. Our work is supported by the Max Planck Innovation Fund (HHR), the DFG SFB 577 (HHR) and the Iranian Molecular Medicine Network (HN).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Moheb, L., Tzschach, A., Garshasbi, M. et al. Identification of a nonsense mutation in the very low-density lipoprotein receptor gene (VLDLR) in an Iranian family with dysequilibrium syndrome. Eur J Hum Genet 16, 270–273 (2008). https://doi.org/10.1038/sj.ejhg.5201967
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201967
Keywords
This article is cited by
-
Systematic review of autosomal recessive ataxias and proposal for a classification
Cerebellum & Ataxias (2017)
-
Very mild features of dysequilibrium syndrome associated with a novel VLDLR missense mutation
neurogenetics (2016)
-
Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion
European Journal of Human Genetics (2013)
-
Cerebellar ataxia, mental retardation and dysequilibrium syndrome 1 (CAMRQ1) caused by an unusual constellation of VLDLR mutation
Journal of Neurology (2013)
-
A missense founder mutation in VLDLR is associated with Dysequilibrium Syndrome without quadrupedal locomotion
BMC Medical Genetics (2012)