Abstract
This study was aimed at identifying the genomic loci linked to exercise participation in males and females. Cross-sectional exercise data of twins and siblings (18–50 years) were used from the Netherlands Twin Registry. The sample consisted of 1432 genotyped sibling pairs from 622 families (1120 sibling pairs were genotyped on all chromosomes). Exercise participation (no/yes, based on a cutoff criterion of four metabolic equivalents and 60 min weekly) was assessed by survey. Genotyping was based on 361 markers and an average marker density of 10.6 cM. Identical by descent status was estimated for a 1 cM grid. A variance components-based sex-limited linkage scan was carried out for exercise participation. The heritability of exercise participation in males was 68.5% and in females 46.3%. The genetic overlap was estimated at 0.32, indicating that partly different genes affect exercise in the two sexes. Suggestive linkage was found in all subjects on chromosome 19p13.3 (LOD=2.18). Although sex differences in linkage effect were not significant, mainly females contributed to the suggestive linkage. The 19p13.3–13.2 region harbors a number of genes related to muscle performance and muscle blood flow, which might affect exercise behavior through exercise ability. Most likely, a large number of genes with each small effects affect exercise participation in males and females. Large collaborative samples are needed to detect these effects.
Similar content being viewed by others
Introduction
Numerous epidemiological and experimental studies have demonstrated the beneficial effects of regular exercise participation on physical and mental health.1, 2, 3 Despite these well-known effects, about 30% of European and North-American populations remain sedentary.4, 5 It is well known that individual differences in exercise behavior can be explained by a combination of both environmental and genetic factors. Reviews of twin and family studies have shown a significant contribution of genetic effects to variation in adolescent and adult exercise participation and (leisure-time) physical activity. Heritability estimates range from 25 to 75%, with the lower estimates found in early adolescence, peak heritability at late adolescence/young adulthood and heritabilities of around 50% in adults.6, 7, 8 At all ages, there is evidence that the genetic factors influencing exercise behavior in males and females are different. Two studies found a higher heritability in male than in female adolescents,6, 9 and a recent study in adults found significant lower correlations in opposite-sex than in same-sex twin pairs in four out of five large data sets from different countries.10 Such a pattern of correlations in first-degree relatives suggests that either different expression patterns of the same genes or different genes play a role in exercise behavior in men and women.
The influence of specific environmental factors on exercise behavior is well researched in the epidemiological literature on determinants of exercise behavior, although causality often still needs to be established.11 In contrast, there is only a handful of molecular genetic studies that identified the actual genetic variants related to exercise behavior. In one study, the dopamine 2 receptor (DRD2) gene was associated with physical activity, sports participation and occupational physical activity in females.12 In a study of adolescent females, the calcium-sensing receptor (CASR) gene was associated with hours spent on physical activities per week.13 In a sample of postmenopausal women, the aromatase (CYP19) gene was associated with physical activity.14 In yet another study, the melanocortin-4 receptor (MC4R) gene was associated with daily physical activity levels in a combined sample of adult men and women.15 Finally, in a study of mild male and female hypertensives, the angiotensin-converting enzyme (ACE) gene was associated with leisure-time physical activity.16
There are two genome-wide linkage studies on physical activity and none on exercise participation.17, 18 In the first study,17 172 male and 223 female adults and their parents from 207 families were genotyped and four physical activity phenotypes were measured. Genotyping was based on 432 markers (average map density 7.06 cM). Three physical activity phenotypes (inactivity, moderate to strenuous physical activity and total daily activity level) were derived from a 3-day activity diary. The fourth physical activity phenotype (time spent on most common physical activity during the past year) was survey-based. For time spent on physical activity, suggestive linkage was found on chromosomes 11p15 and 15q13.3. For the 3-day diary-based physical activity phenotypes, promising evidence was found on chromosome 2p22–p16 (for inactivity) and suggestive linkages were found for different loci on chromosomes 4q28.2, 7p11.2, 9q31.1, 13q22–q31 and 20q13.1.
In a second study,18 1030 children (both boys and girls) and 631 parents from 319 Hispanic-American families were genotyped and phenotyped. Genotyping was based on markers with an average spacing of 10 cM. Daily physical activity was measured using accelerometers. For percentage of awake time spent in sedentary activity, significant linkage was found on chromosome 18q12–q21, where the MC4R gene is located.
There was no overlap in the findings of these linkage studies and, with the exception of MC4R, the genes identified in previous association studies are not located on or nearby the identified regions in the linkage studies. If different genes cause variation in exercise behavior in males and females, as suggested by heritability studies, then ignoring these sex differences might result in a failure to detect the separate genetic effects in males and females. In this paper, we present a sex-limited autosomal linkage scan, carried out in 1570 individuals from 622 families using, on average, 361 markers.
Methods
Subjects
This study was part of an on-going study on lifestyle and health in twin families that are voluntarily registered at the Netherlands Twin Register.19, 20 Since 1991, every 2–3 years, participants received questionnaires on health, lifestyle and personality. Data on exercise participation were collected in each survey in 1991, 1993, 1995, 1997, 2000 and 2002. A cross-sectional data set was created using the most recent data on exercise participation from each family that participated one or more times in the longitudinal study.
Twins and their siblings aged between 18 and 50 years were selected. We excluded twins with unknown zygosity (N=67, note that these twins were not genotyped) and genetically unrelated siblings and half siblings (N=47). The total sample consisted of 4230 families (9408 twins and siblings). A subsample was genotyped and used in the linkage analyses. Genotyping procedures are described below. A detailed overview of the sample characteristics is given in Table 1, Table 2. Zygosity of the same-sex twins was determined by DNA typing for 26.1% of the same-sex twin pairs. For the other same-sex twins, zygosity was based on eight items on physical similarity and the frequency of confusion of the twins by parents, other family members and strangers. Agreement between zygosity based on these items and zygosity based on DNA was 97%.21
Phenotype
Exercise participation was measured with a number of questions. The first question was ‘Do you participate in exercise regularly?’. This question could be answered with Yes or No. If the participants responded affirmative, further information on type, frequency and duration of exercise was gathered. All exercise activities were assigned a metabolic equivalent value, using Ainsworth's Compendium of physical activity.22 A metabolic equivalent score of 1 corresponds to the rate of energy expenditure when at rest (1 kcal/kg/h). In the group of exercisers, the data of frequency and duration of the exercise activities and their metabolic equivalent scores did not follow a normal distribution but were highly skewed. Non-exercisers did not have values on frequency, duration and intensity level, since they are not involved in any exercise activities. Therefore, in keeping with existing epidemiological studies,23 exercise participation was defined as a dichotomous variable, classifying participants as either regular exerciser or non-exerciser. A cutoff criterion of exercising at four metabolic equivalents or more for at least 60 min a week in the recent year was used to classify participants as regular exercisers.
Genotyping
DNA was extracted for a sample of twins, non-twin siblings and their parents from either whole blood or buccal swabs following standard protocols.24, 25 One part of the sample (N=2399 subjects) was genotyped by the Mammalian Genotyping Service in Marshfield for a 400 marker 10 cM genome scan (two batches, screening sets 10 and 16). The other part of the sample (N=985 subjects) was genotyped by the Molecular Epidemiology Section, Leiden University Medical Centre, using the 10 cM Applied Biosystems Human Linkage Set v2.5 MD10 with some additional markers (419 in total). The genotype data from these screens were then combined. Allele calling and binning was equalized between 111 markers that were present in both data sets with the use of 302 overlapping samples. In case there were inconsistencies, the data was set to unknown for the tested markers (binning and allele-calling inconsistencies) and persons (genotyping errors). Sex and zygosity were checked with the marker data. Pedigree relations in the entire samples were checked with the GRR program.26 Errors of Mendelian inheritance were detected with Pedstats.27 Markers and samples were removed if their total error rate was more than 1%; in all other cases, the specific erroneous genotypes were set as unknown. Unlikely, recombinants were detected using Merlin and erroneous genotypes were removed with Pedwipe.27 Identical by descent (IBD) estimation was carried out in this full genotype data set.
There were 1738 monozygotic (MZ) and dizygotic (DZ) twins and non-twin siblings from 622 families who were both genotyped and phenotyped (see also Table 1). For the present linkage analysis, we randomly selected 1 MZ twin from each monozygotic twin pair. Further, siblings were selected per chromosome if they had markers with an average spacing of 18 cM or less. This corresponds to the average spacing of markers if subjects with more than 200 markers genotyped along the genome are selected (ie more than 50%). This resulted in a total sample of 1570 genotyped individuals from 622 families (1432 all possible sibling pairs). The number of sibling pairs per chromosome ranged from 1196 to 1432 (1120 sibling pairs were successfully genotyped on all chromosomes). There were 266 male, 525 female and 641 opposite-sex sibling pairs. The average heterozygosity of autosomal markers was 76%. In the 1120 sibling pairs, the average number of markers genotyped was 361 (201–761) with an average spacing of 10.6 cM. For 1046 pairs, the genotype data came from the Marshfield marker sets; for 171 pairs, the data came from the Leiden marker set. A total of 119 pairs were genotyped in both data sets, and in 22 pairs, the siblings were genotyped in the two different marker sets. For the statistical analyses, the Haldane mapping function was used. All reported values are in Haldane centiMorgans. The marker positions were interpolated via locally weighted linear regression from the National Center for Biotechnology Information build 35.1 physical map positions and the Rutgers genetic map.28, 29
IBD estimation
A sibling pair shares an allele at a specific locus IBD if the allele is inherited from the same ancestor. A sibling pair can share 0, 1 or 2 alleles IBD at a specific locus. Because IBD status is not always known (depending on the availability of genotypic data in the parents, the map density of the markers and the informativeness of the markers), the probability of sharing 0, 1 or 2 alleles IBD needs to be estimated. This was done for a 1 cM grid multipoint scan using the Lander–Green algorithm implemented in Merlin.27 The proportion of alleles shared IBD at a specific position along the genome was estimated as: where p(IBD=1) is the probability that IBD status is 1 and p(IBD=2) is the probability that IBD status is 2.
where p(IBD=1) is the probability that IBD status is 1 and p(IBD=2) is the probability that IBD status is 2.
Linkage analysis
Sibling pair analysis using variance components modeling in Mx30, 31 was used to estimate the heritability and linkage of a putative quantitative trait locus (QTL) to exercise participation in the sample of genotyped sibling pairs. Because exercise participation is a dichotomous variable, we used a threshold liability model, in which the estimated threshold divides the latent standard normal liability distribution into an ‘affected’ and ‘unaffected’ part.32 Estimates of twin correlations and heritabilities in the total sample have been reported in a previous study.10 Before carrying out the linkage analysis, we evaluated whether the prevalences and heritabilities in the genotyped sample were the same as in the total sample. We fitted an AE threshold model with non-scalar sex limitation33 to the data in both the genotyped and non-genotyped sample. In this model, the variance in liability for exercise participation in each sample (which is set at 1) was decomposed into additive genetic (A) and unique environmental (E) components, allowing for both quantitative differences in variance decomposition (ie, different heritabilities in males and females) and qualitative differences (ie, a lower genetic correlation in opposite-sex pairs). We tested for differences in prevalences and variance decomposition in the two samples by constraining the parameters to be equal across samples. The fit of the models was evaluated by use of the log-likelihood ratio test, computing the difference in minus twice the log-likelihood between two nested models. This difference is χ2 distributed. The degrees of freedom equals the difference in degrees of freedom between the two models. An α level of 0.01 was used for this test. A significant χ2 means that the constrained model fits significantly worse than the full model.
Next, we carried out the linkage scan for all positions along the genome, allowing not only for sex limitation in the heritability, but also in the linkage effect.34 The estimate of the proportion of alleles shared IBD in a specific region along the genome (π̂) was used to model the covariance in a sibling pair that is due to the putative locus. If the QTL effect is significant, the predicted (model-based) phenotypic covariance will be largest for sibling pairs sharing all alleles IBD in a specific region along the genome, the covariance will be smallest for sibling pairs sharing no alleles IBD in this region. Figure 1 shows the path model that was used (drawn for opposite-sex (OS) sibling pairs). All path loadings, including the QTL effect on the trait, are allowed to be different in males and females. In full sibling pairs, for reasons of identification, the part of the phenotypic covariance that is not explained by the QTL is usually modeled as a shared background factor, consisting of both common environmental and genetic factors.31 However, because it was known that covariance between sibling pairs is explained by additive genetic factors only,10 we modeled this background factor as an additive genetic factor. The square of the estimate of the QTL effect in the model reflects the proportion of the total variance in liability to exercise participation (fixed at one) that is explained by the QTL. The genetic correlation in the opposite-sex (OS) sibling pairs was freely estimated (but restricted to vary between 0 and 0.5, based on biological plausibility), while the genetic correlation in same-sex sibling pairs was fixed at 0.5. The correlation between QTLs was given by π̂, a value that is specific to each sibling pair (specified as a definition variable in Mx). The threshold was modeled according to the best-fitting model in the heritability analysis, which included an age effect on the threshold.
Path diagram used to model non-scalar sex-limited linkage (shown for an opposite-sex sibling pair). EXM, exercise participation in males; EXF, exercise participation in females; eM, unique environmental path (males); aM, additive genetic path (males); qM, QTL effect (males); eF, unique environmental path (females); aF, additive genetic path (females); qF, QTL effect (females); EM, male-unique environmental factor; EF, female-unique environmental factor; Ar,M, male additive genetic background factor; Ar,F, female additive genetic background factor, Q, quantitative trait locus (QTL); rAr,OS, genetic correlation between additive genetic background factor of an opposite-sex sibling pair, π̂, estimated proportion of alleles shared IBD (identical by descent) at the QTL.
We tested separately the significance of the QTL effects in males and females. To test the significance of the male QTL effect, we compared the fit of the full model with both sex-specific QTL effects with the fit of the model in which the effect of the QTL in males was fixed at 0. Similarly, we evaluated the significance of the female QTL effect. We also evaluated whether the sex differences in QTL effects in males and females were significant, by comparing the model with two QTL effects with the model in which these two effects were equated. We further tested the significance of the QTL effect equated across sex, by comparing the fit of this model with the model in which the QTL parameter was dropped.
Significance of effects was evaluated by use of the likelihood ratio test, from which the log odds ratio (LOD) score can be computed by dividing the obtained χ2 test statistics by 2ln10 (∼4.6). The linkage is considered significant if an LOD larger than 3.6 is observed, corresponding to a genome-wide 5% chance that a significant LOD score due to random fluctuations is found somewhere on the genome, thereby correcting for multiple testing. The linkage is considered suggestive if a LOD score larger than 2.2 is observed, corresponding to the expectation that an LOD score of this magnitude as a result of random fluctuations is found once in a genome scan.35
We also computed the empirical thresholds for suggestive and significant linkage in males and females, by randomly permuting the data sets a 1000 times. Permutations were carried out by randomly assigning the IBD estimates to the sibling pairs, keeping the sibling pairs and IBD structure along the genome intact. Each permuted data set was then analyzed. The empirical thresholds for suggestive linkage in males and in females were computed by obtaining the maximum sex-specific LOD scores for each chromosome out of the 1000 analyses, and determining what sex-specific LOD score occurs a 1000 times out of 22 000. The empirical threshold for suggestive linkage in males was 1.89 and in females 1.91. The thresholds for significant linkage in males and females were computed by recording the maximum sex-specific LOD scores in each linkage scan in one of the permuted data sets, and then determining which sex-specific LOD scores occur 50 out of 1000 times. The empirical threshold for significant linkage in males was 3.22 and in females was 3.21.
Results
The prevalence of exercise participation in the genotyped individuals was not significantly different from the prevalence of exercise in the non-genotyped individuals (χ2=1.44, df=1, P=0.23, see Table 2 for prevalence estimates). There were also no significant differences in heritability of exercise participation in both males and females between the genotyped and non-genotyped sample (χ2=1.67, df=1, P=0.64). In the genotyped sample, the heritability in males was estimated at 68.5% (95% confidence interval (CI): 56.7–82.9%) and in females at 46.3% (95% CI: 26.0–63.6%). The proportion of variance explained by E in males is estimated at 31.5% (95% CI: 15.7–61.8%) and in females at 53.7% (95% CI: 39.2–73.9%). The genetic correlation in opposite-sex pairs is 0.32 (95% CI: 0.08–0.50). These results suggest that no selection occurred with regard to both phenotype and genotype in the genotyped sample, and the results from the linkage analysis can be generalized to the total sample under study. Qualitative sex differences in genetic effects influencing exercise participation were also found in the genotyped sample, which justifies modeling sex-specific QTL effects in the linkage analysis.
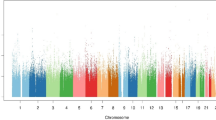
Figure 2 displays the LOD scores for males, females and the combined sample plotted for each chromosome. Tests of sex heterogeneity showed that nowhere along the genome are the sex differences in QTL effect significant, when correcting for multiple testing. Suggestive linkage is found in all subjects on chromosome 19 (maximum LOD=2.18 at 13 cM nearby marker D19S247). It becomes clear from Figure 2 that females contribute more strongly to this LOD score than males (maximum LOD in females=2.87 at 11 and 12 cM, versus 0.83 in males at 9–12 cM). The proportion of variance explained at this QTL is 38.0% (95% CI: 16.8–55.6%). The estimate of the genetic correlation of the additive genetic residual factors in opposite-sex pairs at this locus is zero. Dropping 1 LOD at both sides of the peak, the CI around the peak is 0–28 cM. This region is flanked by markers D19S591 at 19p13.3 and D19S865 at 19p13.2.
Discussion
This study shows suggestive linkage on chromosome 19p.13.3 near marker D19S247 (LOD=2.18), explaining 38.0% of the total variance in liability for exercise participation. The maximum LOD score in females in this region was 2.87 and in males was 0.83. The region on 19p does not coincide with the regions that were found for physical activity levels in the previous linkage studies,17, 18 which could be partly explained by the different definitions of exercise behavior that were used. The region further does not harbor genes that have been related to exercise or physical activity phenotypes in previous association studies.12, 13, 14, 15, 16 Also, no convergence was found to genes on the latest version of the human gene map for performance and health-related fitness phenotypes36 that provides an overview of all genes and QTLs identified through association and linkage studies that have been related to physical performance, physical activity or health-related fitness phenotypes.
A tentative search for genes located under the peak on 19p that are possibly related to exercise participation was made using the Ensembl database.37 We hypothesize that three biological pathways might explain how genes influence exercise behavior. First, genes that influence exercise ability might indirectly influence voluntary engagement in exercise activities. A person's genetic make-up determines whether this person is good at exercise or not, and this innate exercise ability might in turn influence whether a person actually engages in exercise behavior. Second, the engagement in exercise activities might depend on personality traits such as neuroticism, extraversion or sensation seeking, which are all also under genetic control,38, 39, 40, 41 or clinical end points like depression. For example, a linkage study on depressive disorders reported significant linkage of region 19p13.2–13.1 to depressive spectrum disorder.42 Depression, which is two times more prevalent in women, may prevent people to take part in regular exercise. Genes influencing depression might therefore also influence exercise behavior. A third biological pathway by which the influence of genes on exercise behavior might be mediated is through the acute (rewarding) effects of exercise. A person's genetic predisposition to experience more rewarding than aversive acute effects of exercise (eg, large increase in performance or physical fitness, enhanced feelings of well being) may determine whether a person engages and continues to engage in exercise activities. A number of possibly interesting genes are located at region 19p13.3–13.2, all concerning exercise ability through their influence on either muscle performance or muscle blood flow: the muscle integrin-binding protein gene (MIBP), the thyroid receptor-interacting protein 10 gene (TRIP-10), the myosin IE gene (MYO1F), the endothelial differentiation G-protein-coupled receptors 5 and 6 genes (EDG5 and EDG6), the thromboxane A2 receptor gene (TBXA2R) and the calponin-1 gene.
The main limitation of this study is the use of a dichotomy to quantify exercise. It is well known that large samples are needed to detect linkage signals of small effect. The power to detect variance components such as additive genetic QTL variance with ordinal data is even lower than with continuous data.43 We carried out a number of analyses to investigate the power to detect non-sex-specific and sex-specific QTL effects and the power to detect sex differences in QTL effects in the non-scalar sex-limitation model for ordinal data. In the simulations, we assumed heritabilities and a genetic correlation in the opposite-sex pairs that correspond to the values of our real data (heritability males 68%, heritability females 46%, genetic correlation opposite-sex pairs 0.32). With 1440 sibling pairs in total and proportions of male–male, female–female and opposite-sex sibling pairs that correspond to the real data proportions, the power to detect a QTL effect of 25%-explained variance is 0.84 for the male-specific, 0.91 for the female-specific and 0.96 for the non-sex-specific QTL effect. For a QTL explaining 10% of the variance, these values are 0.22, 0.25 and 0.31, respectively. The power to detect a difference of 15%-explained variance between males and females is 0.14 (assuming that the variance in males is 0.05 and in females is 0.20). Similar power results are obtained when different variances are assumed (range between 0 and 20%) but with the same difference of 15%. Thus, the power to detect linkage signals of small effects is rather low and the power to detect sex differences in QTL effects is very low. However, we stress that our sample size is large compared with previous linkage studies on physical activity phenotypes and other linkage studies on complex phenotypes.
Taken together, this study suggests that the substantial heritability of exercise behavior in both males and females cannot be attributed to a few major genes with large effects. Rather, exercise behavior should be considered among the complex, polygenic traits, with in part different genes affecting exercise behavior in males and females. Considering the diversity of the hypothesized biological pathways through which genes might affect exercise behavior, it is likely that a large number of genes with all minor effects account for the heritability of exercise participation. For gene-finding efforts for exercise behavior to be successful, large collaborative samples will be needed to detect and replicate the linkage signals.
References
Salmon P : Effects of physical exercise on anxiety, depression, and sensitivity to stress: a unifying theory. Clin Psychol Rev 2000; 21: 33–61.
Berlin JA, Colditz GA : A meta-analysis of physical activity in the prevention of coronary heart disease. Am J Epidemiol 1990; 132: 612–628.
Farmer ME, Locke BZ, Moscicki EK, Dannenberg AL, Larson DB, Radloff LS : Physical activity and depressive symptoms: the NHANES I epidemiologic follow-up study. Am J Epidemiol 1988; 128: 1340–1351.
Martinez-Gonzalez MA, Martinez JA, Hu FB, Gibney MJ, Kearney J : Physical inactivity, sedentary lifestyle and obesity in the European Union. Int J Obes Relat Metab Disord 1999; 23: 1192–1201.
Caspersen CJ, Pereira MA, Curran KM : Changes in physical activity patterns in the United States, by sex and cross-sectional age. Med Sci Sports Exerc 2000; 32: 1601–1609.
Beunen G, Thomis M : Genetic determinants of sports participation and daily physical activity. Int J Obes Relat Metab Disord 1999; 23: S55–S63.
Stubbe JH, de Geus EJC : Genetics of Exercise Behavior in Handbook of Behavior Genetics. Springer, in press.
Stubbe JH, Boomsma DI, de Geus EJC : Sports participation during adolescence: a shift from environmental to genetic factors. Med Sci Sport Exerc 2005; 37: 563–570.
Maia JAR, Thomis M, Beunen G : Genetic factors in physical activity levels—a twin study. Am J Prev Med 2002; 23: 87–91.
Stubbe JH, Boomsma DI, Vink JM et al: Genetic influences on exercise participation: a comparative study in adult twin samples from seven countries. PLoS ONE 2006; 1: e22. doi:10.1371/journal.pone.0000022.
King AC, Blair SN, Bild DE et al: Determinants of physical activity and interventions in adults. Med Sci Sports Exerc 1992; 24: S221–S236.
Simonen RL, Rankinen T, Perusse L et al: A dopamine D2 receptor gene polymorphism and physical activity in two family studies. Physiol Behav 2003; 78: 751–757.
Lorentzon M, Lorentzon R, Lerner UH, Nordstrom P : Calcium sensing receptor gene polymorphism, circulating calcium concentrations and bone mineral density in healthy adolescent girls. Eur J Endocrinol 2001; 144: 257–261.
Salmen T, Heikkinen AM, Mahonen A et al: Relation of aromatase gene polymorphism and hormone replacement therapy to serum estradiol levels, bone mineral density, and fracture risk in early postmenopausal women. Ann Med 2003; 35: 282–288.
Loos RJF, Rankinen T, Tremblay A, Perusse L, Chagnon Y, Bouchard C : Melanocortin-4 receptor gene and physical activity in the Quebec Family Study. Int J Obes Relat Metab Disord 2005; 29: 420–428.
Winnicki M, Accurso V, Hoffmann M et al: Physical activity and angiotensin-converting enzyme gene polymorphism in mild hypertensives. Am J Med Genet 2004; 125A: 38–44.
Simonen RL, Rankinen T, Perusse L et al: Genome-wide linkage scan for physical activity levels in the Quebec family study. Med Sci Sports Exerc 2003; 35: 1355–1359.
Cai GW, Cole SA, Butte N et al: A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obes Res 2006; 14: 1596–1604.
Boomsma DI, Vink JM, van Beijsterveldt TC et al: Netherlands Twin Register: a focus on longitudinal research. Twin Res 2002; 5: 401–406.
Boomsma DI, de Geus EJC, Vink JM et al: Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet 2007; 9: 849–857.
Willemsen G, Posthuma D, Boomsma DI : Environmental factors determine where the Dutch live: results from the Netherlands Twin Register. Twin Res Hum Genet 2005; 8: 312–317.
Ainsworth BE, Haskell WL, Whitt MC et al: Compendium of physical activities: an update of activity codes and MET intensities. Med Sci Sports Exerc 2000; 32: S498–S516.
Haase A, Steptoe A, Sallis JF, Wardle J : Leisure-time physical activity in university students from 23 countries: associations with health beliefs, risk awareness, and national economic development. Prev Med 2004; 39: 182–190.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Meulenbelt I, Droog S, Trommelen GJM, Boomsma DI, Slagboom PE : High-yield noninvasive human genomic DNA isolation method for genetic-studies in geographically dispersed families and populations. Am J Hum Genet 1995; 57: 1252–1254.
Abecasis GR, Cherny SS, Cookson WOC, Cardon LR : GRR: graphical representation of relationship errors. Bioinformatics 2001; 17: 742–743.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR : Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30: 97–101.
Duffy DL : An integrated genetic map for linkage analysis. Behav Genet 2006; 36: 4–6.
Kong X, Murphy K, Raj T, He C, White PS, Matise TC : A combined linkage-physical map of the human genome. Am J Hum Genet 2004; 75: 1143–1148.
Neale MC, Boker SM, Xie G, Maes HH : Mx: Statistical Modeling, 7th edn. Richmond Virginia: Department of Psychiatry, Virginia Commonwealth University, 2006.
Neale MC : QTL Mapping with Sib-pairs: The Flexibility of Mx in Advances in Twin and Sib-pair Analysis 219–243. London: Greenwich Medical Media Ltd, 2000.
Neale MC, Cardon LR : Methodology for Genetic Studies of Twins and Families. Dordrecht: Kluwer Academic Publishers, 1992.
Medland SE : Alternate parameterization for scalar and non-scalar sex-limitation models in Mx. Twin Res 2004; 7: 299–305.
Medland SE : Parameterization of sex-limited autosomal linkage analysis for Mx. Twin Res Hum Genet 2005; 8: 569–573.
Lander E, Kruglyak L : Genetic dissection of complex traits—guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11: 241–247.
Wolfarth B, Bray MS, Hagberg JM et al: The human gene map for performance and health-related fitness phenotypes: the 2004 update. Med Sci Sports Exerc 2005; 37: 881–903.
Birney E, Andrews D, Caccamo M et al: Ensembl 2006. Nucleic Acids Res 2006; 34: D556–D561.
Stoel RD, de Geus EJC, Boomsma DI : Genetic analysis of sensation seeking with an extended twin design. Behav Genet 2006; 36: 229–237.
Rettew DC, Vink JM, Willemsen G, Doyle A, Hudziak JJ, Boomsma DI : The genetic architecture of neuroticism in 3301 Dutch adolescent twins as a function of age and sex: a study from the Dutch Twin Register. Twin Res Hum Genet 2006; 9: 24–29.
Johnson W, Krueger RF : Genetic and environmental structure of adjectives describing the domains of the Big Five Model of personality: a nationwide US twin study. J Res Pers 2004; 38: 448–472.
Viken RJ, Rose RJ, Kaprio J, Koskenvuo M : A developmental genetic analysis of adult personality: extraversion and neuroticism from 18 to 59 years of age. J Pers Soc Psychol 1994; 66: 722–730.
Zubenko GS, Maher B, Hughes HB et al: Genome-wide linkage survey for genetic loci that influence the development of depressive disorders in families with recurrent, early-onset, major depression. Am J Med Genet Part B Neuropsychiatr Genet 2003; 123B: 1–18.
Neale MC, Eaves LJ, Kendler KS : The power of the classical twin study to resolve variation in threshold traits. Behav Genet 1994; 24: 239–258.
Acknowledgements
This study was supported by the Netherlands Organization for Scientific Research (NWO-904-61-193, NWO-480-04-004 and NWO-575-25-006). We thank the labs of Drs J Weber (Mammalian Genotyping Service, Marshfield) and PE Slagboom (Molecular Epidemiology Section, Leiden University Medical Centre, The Netherlands) for microsatellite genotyping.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
De Moor, M., Posthuma, D., Hottenga, JJ. et al. Genome-wide linkage scan for exercise participation in Dutch sibling pairs. Eur J Hum Genet 15, 1252–1259 (2007). https://doi.org/10.1038/sj.ejhg.5201907
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201907
Keywords
This article is cited by
-
The Genetics of Physical Activity
Current Cardiology Reports (2017)
-
Genetics of Physical Activity and Physical Inactivity in Humans
Behavior Genetics (2012)
-
Effect of the melanocortin-3 receptor Thr6Lys and Val81Ile genetic variants on body composition and substrate oxidation in Chilean obese children
Journal of Physiology and Biochemistry (2012)
-
Determinants of physical activity and exercise in healthy older adults: A systematic review
International Journal of Behavioral Nutrition and Physical Activity (2011)
-
Exercise therapy in Type 2 diabetes
Acta Diabetologica (2009)