Abstract
Limb girdle muscular dystrophy (LGMD) is very common in the Hutterite population of the North American Prairies. We have recently reported the homozygous c.1459G>A mutation in TRIM32 associated with LGMD2H. We have also identified Hutterite patients with LGMD2I, homozygous for the common c.826C>A mutation in FKRP. To date, all Hutterites with LGMD have been shown to be homozygous for either the TRIM32 or FKRP mutation. We now report a Hutterite family in which both parents and five sons were all found to be homozygous for the TRIM32 mutation. The father had slowly progressive proximal muscle weakness, whereas three sons and their mother, all currently asymptomatic, had normal physical examinations. The remaining two sons (7 and 10 years old), presented with mild decrease in stamina, had normal neuromuscular examinations and were found to be homozygous for the FKRP mutation in addition to the TRIM32 mutation. These two boys do not differ in age at or mode of presentation, physical findings, or serum CK levels compared to age-matched individuals affected with LGMD2I alone. This suggests that the effects of these two mutations are not acting synergistically at this time. It remains to be seen whether there will be signs of interaction between these two mutations as the patients get older.
Similar content being viewed by others
Introduction
Limb girdle muscular dystrophies (LGMDs) are a clinically and genetically heterogeneous group of proximal myopathies. To date, they have been mapped to 17 different loci; seven LGMDs have an autosomal dominant mode of inheritance and 10 have an autosomal recessive mode of inheritance.1, 2, 3 Mutations in genes have been identified for three of the dominantly inherited LGMDs and for all of the recessively inherited LGMDs. Among the latter are LGMD2H (OMIM #254110) and LGMD2I (OMIM #607155).
Hutterites are a religious and genetic isolate found throughout the North American plains with a high prevalence of LGMD. LGMD2H is a relatively mild form that was first described in the Manitoba Hutterites in 1976.4 We previously identified c.1459G>A (D487N) in TRIM32 as cosegregating with the disease.5 Subsequently, we found this same mutation causes an infrequently reported muscle disease described as a sarcotubular myopathy (STM), further supporting the fact that D487N is a pathogenic mutation and suggesting that LGMD2H and STM are subtle variants of the same disorder.6 LGMD2I was first described in a large consanguineous Tunisian family and mapped to chromosome region 19q13.3.7 FKRP, encoding fukutin-related protein, was identified as the causative gene in this disorder as well as a congenital muscular dystrophy, MDC1C.8 A mutation in FKRP associated exclusively with the LGMD phenotype, c.826C>A (L276I), is particularly prevalent in Caucasian populations and we determined that this mutation is responsible for the second form of LGMD in Hutterites.9 These two mutations appear to be responsible for all the LGMDs seen in the Hutterites.
The products of these two genes are thought to work through very different mechanisms. TRIM32 is a potential E3 ubiquitin ligase and has antiapoptotic properties.10 Presumably, LGMD2H is a result of a disruption in protein homeostasis leading to myocyte death, a novel mechanism causing muscular dystrophy.5 FKRP is thought to be a glycosyltransferase that functions in the O-linked glycosylation of proteins such as α-dystroglycan, an essential component of the dystrophin-associated glycoprotein complex. In myocytes with mutant FKRP, the glycosylation of α-dystroglycan is insufficient to maintain the link from the contractile apparatus to the extracellular matrix. This makes the cells susceptible to stress-induced damage and may lead to cell death, a common theme in many types of muscular dystrophy.11
Here, we report on two individuals with homozygous mutations at two LGMD loci, LGMD2H and LGMD2I. We present evidence, through the comparison of phenotype and pathology, that homozygosity for both disease mutations does not result in a more severe phenotype at a young age.
Patients and methods
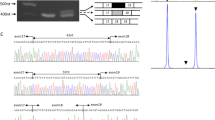
Written informed consent was obtained from all participants. Patients were examined by Dr CR Greenberg and assessed by standard methods. We genotyped individuals for the LGMD mutations and surrounding markers using previously published methods.5, 9 Further confirmation of mutant genotypes was obtained by sequencing both strands using a commercial sequencing service (The Centre for Applied Genomics, Hospital for Sick Children, Toronto, ON, Canada). Haplotypes were constructed manually by minimizing the number of recombinants and assuming no mutation of marker alleles. A muscle biopsy was obtained from the right quadriceps muscle of individual IV:10 (Figure 1) and evaluated using standard methods.
Pedigree and haplotypes. (a) Extended pedigree with the family of interest in the dashed box. Inset legend indicates the meaning of different symbols. NA signifies that the individual was not available for study. Selected individuals were removed and birth order was randomized to maintain anonymity. (b) Haplotypes of chromosome regions 9q32 and 19q13, surrounding the causative mutations of LGMD2H and LGMD2I, respectively (in boxes). Alleles corresponding to the consensus Hutterite disease-associated haplotypes are outlined in yellow or blue, alleles not corresponding are in gray. White haplotypes are the paternal and maternal nondisease-associated haplotypes. Alleles for microsatellite markers are designated by length in bp, single nucleotide polymorphism (SNP) alleles are designated by nucleotide or amino acid (for coding SNPs), and deletions are designated by +/− (not deleted/deleted).
Results and discussion
The prevalence of LGMD in the Hutterite population is much greater than in the general population. Estimates of the world-wide prevalence range from 1/15 00012 to 1/25 000,13 while in the Hutterites we estimate the prevalence to be >1/400. The high frequency of the two alleles causing LGMD in the Hutterites leads to pedigrees that show a pseudodominant pattern of inheritance such as the family reported herein (Figure 1a). This family was ascertained as having LGMD through individual II:1. He was originally described in 1976 by Shokeir and Kobrinsky.4 Most of the remaining individuals were described later and were used to map the LGMD2H locus.14 As there were Hutterite families that were not linked to this locus, another study was undertaken and c.826C>A in FKRP was discovered as the other mutation causing LGMD in this population.8 When the family of individual II:1 was genotyped for c.826C>A, it was found that his wife (II:6) was a carrier as were five of their children (III:3, III:4, III:5, III:6, and III:10).
III:4 had participated in our original LGMD2H family study and was identified to be homozygous for the TRIM32 mutation.5 Although clearly symptomatic he had not himself sought ongoing medical attention. His nuclear family came back to our attention recently due to parental concern of mildly decreased stamina in IV:10 and IV:11 (10 and 7 years). Their past medical histories were unremarkable and both had shown normal growth and development. Detailed clinical examinations of the entire family were performed and findings are shown in Table 1. Both of the boys in question had slender builds with no muscle hypertrophy or atrophy and no joint contractures. In spite of parental concerns, there were no objective signs of muscle weakness, with normal cranial nerves, a negative Gower sign, normal gait, and normal running in the clinical setting. They had normal cardiac and respiratory exams, with echocardiograms showing no evidence of disease. They had elevated serum CK values of 1555 and 1055 U/l, respectively (normal <165 U/l). The other three sons IV:9, IV:12, and IV:13 (11, 6, and 4 years) had similar builds as their brothers and normal physical examinations. Their CK levels were 160, 151, and 213 U/l respectively (normal <165 U/l). Their 33-year-old mother, III:22, was also asymptomatic (CK level 117 U/l; normal 28–110 U/l) and their 40-year-old father, III:4, had moderate slowly progressive proximal muscle weakness and a CK level of 2030 U/l (normal 52-175 U/l).
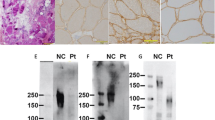
A quadriceps muscle biopsy was performed on individual IV:10 (Figure 2). The muscle showed moderately severe dystrophic changes with increased fiber size variation, scattered necrotic fibers, regenerating fibers, negligible fibrosis, and segmental vacuolation in at least 5% of fibers. The latter is usually only seen in LGMD2H biopsies.6 Vacuoles did not show acid phosphatase activity and did not stain with periodic acid-Schiff, indicating that they were neither autophagic nor associated with glycogen storage. Electron microscopic examination demonstrated clusters of empty vacuoles associated with membranes. In addition, a few otherwise intact fibers had degenerating nuclei with condensed heterochromatin (apoptosis-like) immediately next to normal nuclei (data not shown).
Muscle biopsy from individual IV:10. Panels a and b show photomicrographs of cross-sections of frozen muscle stained with hematoxylin and eosin at × 20 and × 40 magnification, respectively. There is increased fiber size variation and rare fibers have internal nuclei. Panels c and d show photomicrographs of plastic sections in cross (c) and longitudinal (d) orientation taken at × 40 magnification. Small empty appearing vacuoles are present in a subset of fibers.
Genotyping for the two LGMD-causing mutations and surrounding markers was performed. Both parents (III:4 and III:22) and all five sons (IV:9–13) were found to be homozygous for the LGMD2H mutation (Figure 1b). Genotyping of the LGMD2I mutation revealed that both III:4 and III:22 were carriers as were their three asymptomatic sons. The two sons with parental concerns of decreased exercise tolerance (IV:10 and IV:11) were also homozygous for the LGMD2I mutation (Figure 1b).
There is an enormous amount of variation within our LGMD2H cohort (52 patients). We have identified a number of individuals who are asymptomatic even in their fourth decade. Our estimate of the average age of onset is about 24 years and there is a tendency for males to manifest signs and symptoms earlier and have higher serum CK values than females.9 It is not surprising to us that the three boys and their mother, unlike the father, are not showing signs of muscular dystrophy, although we expect the disease to manifest eventually. There is also a large amount of variation within our LGMD2I cohort (24 patients); however, we estimate the average age at onset to be about 12 years (range 2–25 years). This appears to explain why only the two boys homozygous for the LGMD2I mutation are symptomatic at a young age.
Notwithstanding the parental concerns, this family appears to indicate that even a double homozygote does not differ objectively on physical examination from a homozygote for either LGMD2H or 2I at an early age. It appears that the clinical phenotype of these two boys is currently determined by homozygosity for the LGMD2I mutation. However, the biopsy performed on IV:10 shows features distinct for LGMD2H, perhaps even more pronounced than in many other LGMD2H biopsies that we have studied. This may indicate that there is the potential for cumulative effects between the two forms of dystrophy at least from a pathological perspective. We may only see additive clinical effects of the two dystrophies in older patients who are manifesting the effects of both mutations. Final judgement regarding synergy between these two mutations must be reserved until these patients are older.
In summary, this report describes individuals homozygous for mutations in two genes known to be causative of two distinct myopathies. Contiguous gene deletions and cosegregation of deleterious alleles in linkage disequilibrium are well-known causes of multiple genetic disorders in the same person. However, reports describing patients who are homozygous for recessive mutations at independently segregating loci, where the molecular defect has actually been determined, are uncommon.15 Within inbred populations such as the Hutterites where founder effects have increased the frequency of many detrimental alleles, it is not highly unlikely for a situation like this to arise. To our knowledge, this is the first report of two neuromuscular diseases coincident in the same individual(s) and one of only a few that conclusively demonstrates multiple autosomal recessive mutations segregating in the same nuclear family.
References
Kaplan JC, Fontaine B : Neuromuscular disorders: gene location. Neuromuscul Disord 2004; 14: 85–106.
Palenzuela L, Andreu AL, Gamez J et al: A novel autosomal dominant limb-girdle muscular dystrophy (LGMD 1F) maps to 7q32.1–32.2. Neurology 2003; 61: 404–406.
Starling A, Kok F, Passos-Bueno MR et al: A new form of autosomal dominant limb-girdle muscular dystrophy (LGMD1G) with progressive fingers and toes flexion limitation maps to chromosome 4p21. Eur J Hum Genet 2004; 12: 1033–1040.
Shokeir MH, Kobrinsky NL : Autosomal recessive muscular dystrophy in Manitoba Hutterites. Clin Genet 1976; 9: 197–202.
Frosk P, Weiler T, Nylen E et al: Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet 2002; 70: 663–672.
Schoser BG, Frosk P, Engel AG et al: Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann Neurol 2005; 57: 591–595.
Driss A, Amouri R, Ben Hamida C et al: A new locus for autosomal recessive limb-girdle muscular dystrophy in a large consanguineous Tunisian family maps to chromosome 19q13.3. Neuromuscul Disord 2000; 10: 240–246.
Brockington M, Yuva Y, Prandini P et al: Mutations in the fukutin-related protein gene (FKRP) identify limb girdle muscular dystrophy 2I as a milder allelic variant of congenital muscular dystrophy MDC1C. Hum Mol Genet 2001; 10: 2851–2859.
Frosk P, Greenberg CR, Tennese AA et al: The most common mutation in FKRP causing limb girdle muscular dystrophy type 2I (LGMD2I) may have occurred only once and is present in Hutterites and other populations. Hum Mutat 2005; 25: 38–44.
Horn EJ, Albor A, Liu Y et al: RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 2004; 25: 157–167.
Haliloglu G, Topaloglu H : Glycosylation defects in muscular dystrophies. Curr Opin Neurol 2004; 17: 521–527.
Nigro V : Molecular bases of autosomal recessive limb-girdle muscular dystrophies. Acta Myol 2003; 22: 35–42.
Emery AE : Population frequencies of inherited neuromuscular diseases – a world survey. Neuromuscul Disord 1991; 1: 19–29.
Weiler T, Greenberg CR, Zelinski T et al: A gene for autosomal recessive limb-girdle muscular dystrophy in Manitoba Hutterites maps to chromosome region 9q31-q33: evidence for another limb-girdle muscular dystrophy locus. Am J Hum Genet 1998; 63: 140–147.
Jarisch A, Giunta C, Zielen S et al: Sibs affected with both Ehlers-Danlos syndrome type IV and cystic fibrosis. Am J Med Genet 1998; 78: 455–460.
Acknowledgements
We are indebted to the patients and their family members for participating in this study. We thank Ms I Rivera, J Douville, and C Hirst for assistance in obtaining genotypes. This work was funded by the Manitoba Institute of Child Health, the Manitoba Health Research Council, and the Canadian Institutes of Health Research. Dr Del Bigio holds the Canada Research Chair in Developmental Neuropathology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Frosk, P., Del Bigio, M., Wrogemann, K. et al. Hutterite brothers both affected with two forms of limb girdle muscular dystrophy: LGMD2H and LGMD2I. Eur J Hum Genet 13, 978–982 (2005). https://doi.org/10.1038/sj.ejhg.5201436
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201436
Keywords
This article is cited by
-
Thin is required for cell death in the Drosophila abdominal muscles by targeting DIAP1
Cell Death & Disease (2018)
-
The ubiquitin ligase tripartite-motif-protein 32 is induced in Duchenne muscular dystrophy
Laboratory Investigation (2016)