Abstract
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disease, characterized by degeneration of the anterior horn cells of the spinal cord. SMA presents with a highly variable phenotype ranging from very severe to mild (type I–III). No cure for SMA is available at present. All forms of SMA are caused by homozygous loss of the functional survival motor neuron (SMN1) gene. However, all patients have one or more copies of the SMN2 gene, nearly identical to SMN1. Both genes encode the SMN protein but the level produced by SMN2 is insufficient to protect from disease. Increasing SMN2 gene expression could be of considerable therapeutic importance. The aim of this study was to assess whether SMN2 gene expression can be increased by 4-phenylbutyrate (PBA). Fibroblast cell cultures from 16 SMA patients affected by different clinical severities were treated with PBA, and full-length SMN2 transcripts were measured by real-time PCR. In all cell cultures, except one, PBA treatment caused an increase in full-length SMN2 transcripts, ranging from 50 to 160% in type I and from 80 to 400% in type II and III cultures. PBA was found also effective in enhancing SMN protein levels and the number of SMN-containing nuclear structures (gems). These data show that SMN expression is considerably increased by PBA, and suggest that the compound, owing also to its favorable pharmacological properties, could be a good candidate for the treatment of SMA.
Similar content being viewed by others
Introduction
Proximal spinal muscular atrophies (SMAs) are a group of clinically heterogeneous motor neuron disorders characterized by the degeneration of the anterior horn cells of the spinal cord. On the basis of age of onset and severity of the clinical course, childhood-onset SMA can be classified into three forms (type I–III). Type I, or Werdnig–Hoffmann disease, is the most severe form and is a leading cause of infant mortality. All forms of SMA are caused by homozygous loss of the survival motor neuron (SMN1) gene due to gene deletion, conversion or mutation.1 SMN1 and a nearly identical copy, SMN2, are located in a duplicated inverted region at 5q13.1 Both genes encode the SMN protein. SMN1 can be distinguished from SMN2 by two base changes in exons 7 and 8; in particular, the C–T transition in exon 7 affects the activity of an exonic splicing enhancer in SMN2 such that the two genes generate SMN isoforms in different proportions.2,3 SMN1 produces primarily full-length (fl) transcripts, whereas SMN2 produces primarily an isoform lacking exon 7 (Δ7) that is presumably unstable.2 Patients have at least one, usually two to four SMN2 genes, and higher copy numbers are generally associated with milder phenotypes.4,5,6,7 The SMN protein is expressed in most tissues and is localized in the cytoplasm and in the nucleus, where it appears concentrated in dot-like structures known as gems.8 The level of SMN protein and the number of gems are markedly reduced in SMA patients. SMN protein levels and gem numbers inversely correlate with the phenotypic severity.9,10,11 This correlation suggests that an increase in functional SMN protein could be beneficial for the patients.
At present, no cure for SMA is available. Three clinical trials for SMA patients have been performed. Gabapentin, a likely neuroprotective agent, did not lead to differences in the outcome measure,12 whereas, in pilot trials using thyrotropin-releasing hormone and albuterol, improvement of muscle strength in a small number of type II and III patients has been reported.13,14 None of these drugs appears to act on the molecular defect. Essentially two strategies to increase SMN2 fl transcripts can be envisaged: one leading to the upregulation of the SMN2 genes and the other resulting in the modification of the SMN2 mRNA splicing pattern. Interferon-stimulated gene expression inducing SMN mRNA has been reported.15 Moreover, aclarubicin, a tetracyclin derivative, has been shown to increase fl SMN production by modifying the isoform ratio.16 However, the side effects of interferon treatment and the toxicity of aclarubicin advice against the use of these drugs for therapy of young SMA patients. Recently, Chang et al17 have reported that treatment with sodium butyrate (BA) of SMA-like mice resulted in increased expression of the SMN protein in spinal motor neurons. Independently, we found that both BA18 and its derivative, the aromatic fatty acid phenylbutyrate (PBA),19 increase SMN2 expression in fibroblast cell lines from SMA patients. PBA, an FDA-approved drug, has been used for several years for the treatment of urea cycle disorders in young patients, and is well tolerated.20 We show here that PBA enhances SMN expression in fibroblast cultures from SMA patients.
Materials and methods
Cell culture and treatment
Human fibroblasts were isolated from skin biopsies obtained from four type I, six type II and six type III SMA patients. Informed consent was obtained from all patients or their parents. For all patients, molecular analysis confirmed the homozygous absence of SMN1 exon 7. Fibroblasts were used within eight passages and plated 1 day before the experiment at a density of 6000 cells/cm2. Cells were routinely maintained in Dulbecco's modified Eagle medium (DMEM) with 4.5% glucose, supplemented with 10% heat inactivated fetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin (all from Gibco BRL, Invitrogen Ltd, Paisley, Scottland, UK). A stock solution of 500 mM 4-phenylbutyric acid (PBA, Fluka) was prepared in ethanol and diluted to final concentration in the culture medium immediately prior to use.
Quantitative real-time PCR of SMN2 gene transcripts
Total RNA was extracted from treated and untreated cell cultures using TRIzol reagent (Invitrogen) or Tri-reagent (Sigma Aldrich). RNA concentrations were determined using the GeneQuant II spectrophotometer (Pharmacia) and 2 μg RNA were used for first-strand cDNA synthesis, as described elsewhere.11 Real time RT-PCR was performed using the ABI PRISM 7700 Sequence Detector System (Applied Biosystems, Foster City, CA, USA). SMN fl mRNA was amplified using a forward primer in SMN exon 6 (5′-GCT GAT GCT TTG GGA AGT ATG TTA-3′) and a reverse primer in SMN exon 7 (5′-CAC CTT CCT TCT TTT TGA TTT TGT C-3′); the sequence of the Taqman probe, localized in exon 6, was 5′FAM-TTT CAT GGT ACA TGA GTG GCT ATC ATA CTG GCT ATT AT-TAMRA 3′. SMNΔ7 isoform was amplified using a forward primer spanning the exon 5 and 6 junction (5′-TGG ACC ACC AAT AAT TCC CC-3′) and a reverse primer spanning the exon 6 and 8 junction (5′-ATG CCA GCA TTT CCA TAT AAT AGC C-3′); the Taqman probe annealed to a sequence in exon 6 (5′FAM-ACC ACC TCC CAT ATG TCC AGA TTC TCT TGA TG-TAMRA 3′). PCR reaction was performed in a total volume of 12.5 μl containing 10 ng cDNA, 1.1 pmol of each primer, 1.25 pmol of Taqman probe and half volume of 2 × Taqman Universal PCR Master Mix (Applied Biosystems). The relative amount of transcripts was calculated by using the threshold cycle (Ct) method, according to the manufacturer's instructions, comparing SMN versus glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Applied Biosystems) transcripts. The results were normalized in each culture as a ratio between relative amount of transcripts in treated versus untreated samples. Real-time PCR reactions of each sample were performed in triplicate and experiments were repeated at least three times.
Flow cytometry
The proportion of cells in G1, S and G2 was determined by flow-cytometric analysis of DNA content (EPICS XL-MCL Flow cytometer, Coulter Electonics). Cells treated with 2 mM PBA for 48 h and controls were trypsinized and pelleted. Following resuspension in PBS, cells (2 × 106 cells/ml) were fixed in 80% ethanol, washed and incubated with 0.5 ml PBS and 0.5 ml DNA-Prep stain containing RNase (4KU/ml) and propidium iodide (50 mg/ml), NP-40 (0.6%), at room temperature for 30 min. DNA histograms were analyzed using Multicycle AV software (Phoenix) to evaluate cell cycle compartments.
Western blot analysis
Fibroblasts were plated at a density of 5 × 105 cells/75 cm2 flasks and incubated with 2 mM PBA for 16 and 24 h. Proteins were extracted from treated and untreated cells and Western blot was performed as described elsewhere.11 The antibodies were anti-SMN (dilution 1/5000, Transduction Laboratories), anti-β-tubulin (dilution 1/12 000) and anti-actin polyclonal antibody (dilution 1/2000, Sigma-Aldrich), anti-mouse horseradish peroxidase-conjugated (dilution 1/2000) and anti-rabbit horseradish peroxidase-conjugated (dilution 1/2000, Amersham Pharmacia Biosciences). Western blot quantification was performed as described;11 SMN–actin–tubulin ratios were determined and normalized to untreated samples. For each sample, at least three different Western blots were performed.
Immunofluorescence analysis
Immunolocalization of SMN protein was performed, as described elsewhere, on untreated and treated cells for 16–24 h.11. The primary anti-SMN antibody (Transduction Laboratories) was diluted 1/500; the secondary antibody (goat anti-mouse FITC-conjugated, Jackson Laboratories) was diluted 1/100. All experiments were performed at least three times. The gem number was evaluated by screening of at least 100 cells/slide by two different investigators, blinded to the treated/untreated status of the cultures.
SMN2 copy number assessment
Estimation of the number of SMN2 genes was performed by real-time PCR. Briefly, allelic discrimination assay with two probes (SMN1 and SMN2) was performed. SMN2 Ct values were normalized to those of the telomerase gene. SMN2 copy number was calculated using as reference, samples of patients with known SMN2 copy number, previously determined by FISH.5 The method will be described in detail elsewhere (manuscript in preparation).
Statistical analysis
All data are expressed as mean±SD. For real-time PCR and Western blot experiments, the ANOVA test was used to evaluate the changes in transcript and protein levels. For gem number studies, the variance between the means was calculated by using a nonparametric test (Friedman test, SPSS, program version 8.0). A P-value less than 0.05 was considered statistically significant.
Results
Effect of PBA on SMN2 gene expression
In a preliminary study of the effect of BA and PBA on SMN2 gene expression in primary fibroblast cell cultures from two SMA patients, we observed an increase in fl transcripts following treatment of the cultures by both drugs.18,19 Given the more favorable pharmacological characteristics of PBA compared to BA, including a longer half-life in vivo,21 we decided to use PBA for further experiments.
A bank of 16 fibroblast cell lines was established from patients affected by various clinical severities, and the effect of PBA on SMN2 expression in the cell cultures was assessed by real-time PCR (Table 1). First, to determine the most effective concentration of PBA on SMN expression, three cell lines (one from each SMA type) were exposed to increasing concentrations of PBA. The levels of SMN2 fl transcripts were assessed after 4 and 16 h of treatment. For the type I culture, similar results were obtained with 1 and 2 mM concentrations of PBA, whereas for the type II and III cultures maximum response was obtained with 2 mM concentration of PBA (Figure 1a). Therefore, we chose to use 2 mM concentration of PBA for further experiments. Complete absence of response in all cultures analyzed was observed with 5 mM PBA, probably due to the toxic effect of the compound at this concentration.
Effect of 4-PBA on SMN2 gene expression in primary fibroblast cultures from SMA patients. Percent increase in transcripts in PBA-treated relative to untreated cultures is shown. (a) Effect of different concentrations of PBA on SMN mRNA synthesis. (b) Increase in SMN2 fl transcripts for each cell line at the treatment time where maximum increase was observed. (c) Effect of repetitive administration of PBA (every 4 h) compared to that of a single dose. (d) Measurement of fl and Δ7 SMN2 transcripts in three cell cultures. Values are mean of at least three real-time PCR experiments performed in triplicate. The error bars indicate SD.
PBA, similar to BA, is known to alter cell proliferation.22,23 To determine the effect of PBA on cell cycle progression in our experimental conditions, two different cultures were treated with 2 mM PBA for 48 h and then analyzed by flow cytometry. A higher percentage of cells in the S phase was found in PBA-treated cells compared to untreated cultures (42–48 versus 28–25%, respectively), which does not support an inhibitory effect on cell cycle progression by the compound (data not shown).
All 16 cultures were then treated with a single dose of 2 mM PBA for different time intervals (4, 8, 16 and 24 h) and SMN2 fl transcripts were measured. A marked variability of response to PBA treatment was observed for both the single cultures at the different treatment time intervals and among the different cell lines. As shown in Figure 1b, all cell cultures from type II and III patients, except one, showed a maximum increase in SMN2 fl transcripts of at least 80% up to 400%, whereas in the four cultures from type I patients the maximum increase ranged from approximately 50 to 160%. The mean values±SD of percent increase in fl transcripts (statistically significant, P=0.01) determined at the four treatment intervals for each culture are indicated in Table 1. In none of the experiments, considerable differences in GAPDH Ct values between treated and untreated cultures were detected, suggesting that PBA has no effect on GAPDH expression and that GAPDH is therefore an appropriate control in the real-time PCR experiments.
To determine whether repetitive administration of PBA could enhance the response to PBA treatment in a low responding cell culture, cells from a type I patient (patient 4) were treated with 2 mM PBA every 4 h. After 24 h, a 100% increase in transcripts was found. Considering that administration of a single dose of PBA to the same cell culture resulted in a maximum increase of about 80% after 8 h, which dropped to about 25% after 24 h, these data suggest that frequent re-exposure to the compound could prevent the decline of transcript level (Figure 1c).
To investigate whether PBA influences alternative splicing of exon 7, we studied the effect of PBA on the synthesis of both SMN2 fl and Δ7 transcripts in three cell cultures. We found that both isoforms were increased to a similar extent at some time points, suggesting the upregulation of the SMN2 genes, whereas in other experiments minor or no increase in Δ7 transcripts was observed, which could be consistent with a possible effect on alternative splicing (Figure 1d).
Effect of PBA on SMN protein expression
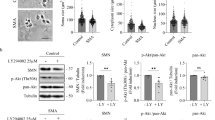
To determine whether PBA treatment has an effect also on SMN protein level, both Western blot and immunofluorescence analysis were performed in some of the cell lines. For Western blot, three cell cultures (one from each SMA type) were treated for 16 and 24 h. Protein levels were quantified, compared to those in untreated cells, and normalized to two loading controls. A marked, although not statistically significant, increase in SMN protein was found, ranging from 36 to 57% after 16 h treatment. Slightly higher protein levels were measured after 24 h incubation time, reaching 46, 42 and 89% increases in the type I, II and III cell cultures, respectively (Figure 2a, b). Both tubulin and actin were used as loading controls to reduce the probability that SMN protein determination could be influenced by an effect of PBA on the expression of the control proteins. No remarkable differences in the ratio between these proteins were observed, suggesting that PBA either does not modulate their expression or has a similar effect on both proteins (data not shown).
Effect of PBA on SMN protein expression. (a) Representative Western blot experiments of SMN protein in SMA fibroblasts. (b) An increase in SMN protein was observed in three cell lines after a 16–24 h PBA treatment. Protein levels were quantified by densitometric analysis and normalized to β-tubulin and actin, used as loading controls. Values are means of at least three different blots. (c) Percent of nuclei with gems and (d) number of gems/100 nuclei in PBA-treated and untreated cell lines observed by immunofluorescence analysis. Mean values are shown as determined from at least three experiments for each cell line. The error bars indicate SD; UN: untreated.
Immunolocalization of the nuclear SMN protein was performed on four cell cultures (two type I, one type II and one type III). We observed a statistically significant increase in both the number of nuclei with gems (P=0.05) and the number of gems/100 nuclei (P=0.04) in all PBA-treated cell cultures compared to the untreated lines (Figure 2c, d). There was a particularly high increase in the total number of gems/100 nuclei in the cultures from the two type I patients (3.5- and 2.9-fold at 24 h).
Response to PBA treatment in relation to the number of SMN2 genes
To investigate the possibility that the response to PBA treatment of the single cultures could be related to the number of SMN2 genes, we determined the SMN2 copy number for each patient (Table 1). Type I patients had one or two genes, whereas the type II and III patients had three or four copies. These data confirm an inverse correlation between disease severity and number of SMN2 genes. Generally, a higher response to PBA treatment was found in patients with more than two SMN2 genes, but there was no absolute correlation between the extent of response and gene number.
Discussion
SMA presents with a highly variable phenotype. Disease severity is modulated by the amount of functional SMN protein produced from the SMN2 gene locus.9 Increasing SMN2 gene expression could be of invaluable therapeutic importance. PBA has been shown to stimulate or modulate the expression of a variety of genes including γ-globin genes in patients with β-hemoglobinopathies.24 The aim of our study was to determine whether PBA is effective in increasing the expression of the SMN2 genes. Our efforts focused in particular on the evaluation of SMN2 transcripts in PBA-treated SMA cell lines by quantitative real-time PCR. Here, we show that PBA causes an 80–400% increase in SMN2 fl transcripts in all but one cell line from type II–III SMA patients relative to the untreated cultures. The compound was less effective in inducing SMN2 expression in cultures from severely affected patients. However, repetitive administration of the compound to one of the low-responding cultures led to an improvement in the response to PBA exposure.
Some cell lines were also subjected to protein studies. Western blot analysis showed that SMN protein levels were enhanced, albeit more moderately as compared to the observed transcript levels, which could reflect the more limited sensitivity of protein quantification by Western blotting compared to transcript quantification by real-time PCR and/or differences in gene and protein activation at the respective treatment times. PBA was also found to determine a marked increase in the number of gems; this effect was particularly evident in the type I cell lines. A relatively high number of nuclei with two or more gems was found after treatment; such nuclei are particularly rare in cells from SMA patients.10,11 Interestingly, PBA treatment for 24 h leads to an average increase in the number of nuclei with gems and gems/100 nuclei, which roughly corresponds to 50% of the levels previously detected in controls.11 The appearance of gems in the nuclei after PBA exposure may reflect the assembly of a higher number of SMN protein complexes due to an increase in SMN fl protein.
The causes of the variability in response to PBA treatment among the different cultures are unknown. It is possible that the response to PBA is related to the number of SMN2 genes. In this case, a stronger response in cell cultures from patients with a higher copy number of SMN2 genes would be expected. SMA type II and III patients have generally more SMN2 genes than type I patients,4,5,6,7 which was confirmed in the present patients cohort. Our observation of a better response to treatment in most cultures from the mildly affected patients supports the above hypothesis. However, there were differences in the extent of response to PBA in some cultures from patients with identical SMN2 gene number. For example, three SMA type I patients investigated had two copies of SMN2, but the cell line from one of these patients showed a two- to three-fold higher increase in transcripts compared to the other two. The mode of action of PBA on the SMN2 genes is not known, but one possible hypothesis to explain this finding is that PBA, an inhibitor of histone deacetylases, modulates transcriptional activity by epigenetic modification of chromatin at the SMN2 locus and that the extent of activation could vary for the single genes.
Chang et al reported that BA influences SMN2 gene expression by inducing SR proteins that modify the splicing pattern of exon 7 of SMN2 transcripts. It could thus be possible that PBA also acts similarly. Our observation of an increase in both fl and Δ7 transcripts in most but not all experiments suggests that enhancement in SMN2 expression may be ascribed at least in part to upregulation of the SMN2 genes, but a reduction of alternative splicing also seems to contribute to the response. Unknown factors such as modifier genes may also contribute to the variability in the increase in both isoforms. Further experiments are necessary to unravel the mode of action of PBA on SMN gene expression.
BA and PBA have antiproliferative effects on the cell cycle, which is exploited in cancer cell lines and clinical trials of patients with cancer.25,26 Our observation of a higher percentage of cells in the S phase which differs from the growth arrest at the G1 phase reported for a cystic fibrosis cell line treated with 5 mM PBA23 may be due to the different experimental conditions.
Clinical studies will be needed to determine whether PBA is also able to enhance SMN expression in vivo. In the plasma of patients with urea cycle disorders treated with 600 mg/kg/day, a PBA concentration of up to 2 mM has been detected, which corresponds to the concentration resulting in the best response in our in vitro experiments.27 PBA is relatively stable with a plasmatic half-life of 0.8–1 h;21 our observation of a better response after repetitive administration of PBA compared to that of a single dose in one of the low-responding cell lines may reflect a beneficial effect on SMN expression of minor fluctuations in PBA concentration in vitro. Furthermore, it has been shown that PBA is able to lower very-long-chain fatty acid levels in the brain of mice with X-linked adrenoleukodystrophy, suggesting that PBA can cross the blood–brain barrier.28 More recently, evidence was given that PBA penetrates well into the CSF after i.v. administration in nonhuman primates.29 These data raise the possibility that PBA could stimulate SMN expression in motor neurons.
In conclusion, our data suggest that PBA could be a good candidate for a pharmacological treatment of SMA patients. Clinical studies are necessary to establish whether PBA enhances SMN gene expression also in vivo and is able to improve muscle strength in SMA patients.
References
Lefebvre S, Bürglen L, Reboullet S et al: Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–165.
Lorson CL, Hahnen E, Androphy EJ, Wirth B : A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA 1999; 96: 6307–6311.
Monani UR, Lorson CL, Parsons DW et al: A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN. Hum Mol Genet 1999; 8: 1177–1183.
McAndrew PE, Parsons DW, Simard LR et al: Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNt and SMNc gene copy number. Am J Hum Genet 1997; 69: 1411–1422.
Vitali T, Sossi V, Tiziano F et al: Detection of the survival motor neuron (SMN) genes by FISH: further evidence for a role of SMN2 in the modulation of disease severity in SMA patients. Hum Mol Genet 1999; 8: 2525–2532.
Brahe C : Copies of the survival motor neuron gene in spinal muscular atrophy: the more, the better. Neuromusc Disord 2000; 10: 274–275.
Feldkötter M, Schwarzer V, Wirth R, Wienker TI, Wirth B : Quantitative analysis of SMN1 and SMN2 based on real-time LightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 2002; 70: 358–368.
Liu Q, Dreyfuss G : A novel nuclear structure containing the survival of motor neurons protein. EMBO J 1996; 15: 3555–3565.
Lefebvre S, Burlet P, Liu Q et al: Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 1997; 16: 265–269.
Coovert DD, Le TT, McAndrew PE et al: The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 1997; 6: 1205–1214.
Patrizi AL, Tiziano F, Zappata S, Donati A, Neri G, Brahe C : SMN protein analysis in fibroblast, amniocyte, and CVS cultures from spinal muscular atrophy patients and its relevance for diagnosis. Eur J Hum Genet 1999; 7: 301–309.
Miller RG, Moore DH, Dronsky V et al: A placebo-controlled trial of gabapentin in spinal muscular atrophy. J Neurol Sci 2001; 191: 127–131.
Tzeng AC, Cheng J, Fryczynski H et al: A study of thyrotropin-releasing hormone for the treatment of spinal muscular atrophy: a preliminary report. Am J Phys Med Rehabil 2000; 79: 435–440.
Kinali M, Mercuri E, Main M et al: Pilot trial of albuterol in spinal muscular atrophy. Neurology 2002; 59: 609–610.
Baron-Delage S, Abadie A, Echaniz-Laguna A, Melki J, Beretta L : Interferons and IRF-1 induce expression of the survival motor neuron (SMN) genes. Mol Med 2000; 6: 957–968.
Andreassi C, Jarecki J, Zhou J et al: Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 2001; 10: 2841–2849.
Chang J-G, Hsieh-Li H-M, Jong Y-J, Wang NM, Tsai C-H, Li H : Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci USA 2001; 98: 9808–9813.
Vitali T, Tiziano F, Neri G, Brahe C : SMN2 gene activation by sodium butyrate and acetyl-L-carnitine in cell cultures from SMA patients; Abstract, Fifth Annual North American/International Spinal Muscular Atrophy Research and Clinical Meeting; June 22–24, 2001; Chicago.
Tiziano FD, Andreassi C, Angelozzi C et al: Upregulation of SMN2 gene expression by 4-phenylbutyrate: relevance for a therapeutic approach to spinal muscular atrophy. Am J Hum Genet 2002; 71 (Suppl): A2445.
Maestri NE, Brusilow SW, Clissold DB, Bassett S : Long term treatment of girls with ornithine transcarbamylase deficiency. N Engl J Med 1996; 335: 855–859.
Gilbert J, Baker SD, Bowling MK et al: A phase I dose escalation and bioavailability study of oral sodium phenylbutyrate in patients with refractory solid tumor malignancies. Clin Cancer Res 2001; 7: 2292–2300.
Davis T, Kennedy C, Chiew Y-E, Clarke CL, deFazio A : Histone deacetylase inhibitors decrease proliferation and modulate cell cycle gene expression in normal mammary epithelial cells. Clin Cancer Res 2000; 6: 4334–4342.
McGrath-Morrow SA, Stahl JL : G1 phase growth arrest and induction of p21waf1/Cip/Sdi1 in IB3-1 cells treated with 4-sodium phenylbutyrate. J Pharmacol Exp Therap 2000; 294: 941–947.
Dover GJ, Brusilow S, Charache S : Induction of fetal hemoglobin production in subjects with sickle cell anemia by oral sodium pehylbutyrate. Blood 1994; 84: 339–343.
Gore SD, Samid D, Weng L-J : Impact of the putative differentiating agents sodium phenylbutyrate and sodium phenylacetate on proliferation, differentiation, and apoptosis of primary neoplastic myeloid cells. Clin Cancer Res 1997; 3: 1755–1762.
Carducci MA, Nelson JB, Chantack KM et al: Phenylbutyrate induces apoptosis in human prostate cancer and is more potent than phenylacetate. Clin Cancer Res 1996; 2: 379–387.
Brusilow SW, Maestri NE : Urea cycle disorders: diagnosis, pathophysiology and therapy. Adv Pediat 1996; 23: 127–170.
Kemp S, Wei H-M, Lu J-F et al: Gene redundancy of pharmacological gene therapy: implications for X-linked adrenoleukodystrophy. Nat Med 1998; 4: 1261–1268.
Berg S, Serabe B, Aleksic A et al: Pharmacokinetics and cerebrospinal fluid penetration of phenylacetate and phenylbutyrate in the nonhuman primate. Cancer Chemother Pharmacol 2001; 47: 385–390.
Acknowledgements
We thank the families for their kind cooperation and are grateful to Dr A Valenza for his help in densitometric scanning of the autoradiograms and to Professor G D'Abramo for his help in statistical analysis. The study was supported by grants from the Associations ASAMSI and Italian Families of SMA, Telethon and MURST 60%.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Andreassi, C., Angelozzi, C., Tiziano, F. et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet 12, 59–65 (2004). https://doi.org/10.1038/sj.ejhg.5201102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201102
Keywords
This article is cited by
-
Evaluation of the orally bioavailable 4-phenylbutyrate-tethered trichostatin A analogue AR42 in models of spinal muscular atrophy
Scientific Reports (2023)
-
Therapeutic potential of endoplasmic reticulum stress inhibitors in the treatment of diabetic peripheral neuropathy
Metabolic Brain Disease (2023)
-
Applicability of Histone Deacetylase Inhibition for the Treatment of Spinal Muscular Atrophy
Neurotherapeutics (2013)
-
Spinal Muscular Atrophy and the Antiapoptotic Role of Survival of Motor Neuron (SMN) Protein
Molecular Neurobiology (2013)
-
Clinical and molecular cross-sectional study of a cohort of adult type III spinal muscular atrophy patients: clues from a biomarker study
European Journal of Human Genetics (2013)