Abstract
Spondyloepiphyseal dsplasia tarda (SEDL) is a radiologically distinct, X-chromosome linked primary skeletal dysplasia characterised by disproportionate short-trunked short stature, dysplasia of the large joints (hip) and flattened thoracic and lumber vertebral bodies. Molecular basis for SEDL has been elucidated by the identification of various mutations (currently >30) in the SEDL gene from Xp22 region. The function of the SEDL protein is not known although it is speculated that it may participate in the ER-to-Golgi transport as part of a novel highly conserved multiprotein TRAPP complex.
Similar content being viewed by others
Clinical definition
Spondyloepiphyseal dysplasia tarda (SEDL) is a well-defined, X-linked primary skeletal dysplasia that predominantly affects the spinal vertebral bodies and epiphyses during skeletal growth.1 The condition is not evident at birth, the usual age of presentation being after the first decade of life. Disproportionate (short-trunked) short stature in a male, with or without back pain, is the common presenting feature. Other characteristic clinical features include a broad chest with mild sternal protrusion and limitation of joint motion at the hips and elbows.2 Craniofacial appearance, vision, hearing and intelligence are unaffected in SEDL and there are no consistently associated extraskeletal anomalies. The radiographic manifestations of SEDL are diagnostic, clearly distinguishing it from other conditions that it has most recently been grouped together within the current classification3 (see Table 1). The most characteristic of these is seen in the lateral view of the thoraco-lumbar spine; comprising generalised platyspondyly, narrowing of intervertebral disc spaces, and pathognomonic superior and inferior ‘humps’ involving the posterior two-thirds of the flattened vertebral bodies2 (see Figure 1). These changes are best appreciated in late childhood and adolescence, and may become superimposed by secondary arthritic changes in later decades. The major potential medical complication of the disorder is premature arthritis, predominantly affecting the spine and hip joints.1 Hip joint disease may be severe, necessitating replacement in early adult life.1 There is a wide range of inter- and intrafamilial variability among affected males in regard to severity of disease. Females have also been reported with classic radiographic features of SEDL,4 but this occurrence is rare, if not unique, and in such females mutations in the SEDL gene are yet to be identified.
Diagnosis
Diagnosis is suggested by the presence of disproportionate (short-trunk) short stature in a male (typically aged 8–15 years) and confirmed by the characteristic vertebral morphology on a lateral radiograph of the thoraco-lumbar spine. Full radiographic survey of the skeleton may also reveal small and irregular epiphyses in childhood and evidence of osteoarthritic change in later life. The most important diagnostic ‘tools’ are a three-generation family history (suggestive of X-linked inheritance) and radiographic skeletal survey. Diagnosis can now also be verified by the finding of a mutation in the SEDL gene.
Differential diagnosis
SEDL is readily differentiated from the other currently classified (Table 1) spondyloepiphyseal dysplasias by age of onset, pedigree analysis and the above radiographic findings on lateral spinal radiographs. There are other late-onset spondyloepiphyseal dysplasias that are not well characterised, but none of these have the typical vertebral morphology change seen in SEDL, and many have other differentiating clinical findings. Perhaps the most common reason for not finding a SEDL mutation in an ‘affected’ male is misdiagnosis.
Gene
The SEDL gene was identified by the screening of positional candidate genes from within a minimal ∼170 kb interval in Xp22 refined by linkage mapping on two families segregating SEDL.5 It is a small gene, which escapes X-inactivation, composed of six exons spanning 20 kb of genomic DNA. Ubiquitously expressed SEDL mRNA is about 2.8 kb in size. It is alternatively spliced involving exons 2, 4 and 6. SEDL coding region of 420 bp is split among exons 3–6. Exons 1 and 2 are noncoding. Exon 2 contains a MER20 repetitive sequence and there are four Alu repeats within the 3′ untranslated region. SEDL gene is a highly conserved gene with orthologs identified in yeast, fly and vertebrates.5
Pseudogenes
SEDL gene has at least seven pseudogenes in the human genome: one SEDLP1 on chromosome 19q13.4; one SEDLP2 on chromosome 8q13.3; and five SEDLP3-SEDLP7, pseudogenes on chromosome Yq11.23.6 While SEDLP1 and SEDLP2 are processed pseudogenes generated by retrotransposition, the Y-chromosome-bound pseudogenes arose by duplication. Interestingly, the chromosome 19 pseudogene, SEDLP1 is transcribed (ubiquitously) from a novel promoter and encodes for a protein, which would be 100% identical to that of the SEDL gene. It still remains to be determined whether the SEDLP1 mRNA of 0.75 kb is translated. There are only six silent nucleotide differences between the open reading frame of the SEDL gene and the SEDLP1 pseudogene. Other SEDL pseudogenes are not transcribed.6
Genetic heterogeneity
Although X-linked SEDL can be distinguished from the other currently classified spondyloepiphyseal dysplasias by typical radiographic findings, not all such patients have SEDL gene mutations identified (15% or six cases with clinically defined isolated SEDL were negative when tested for SEDL gene mutations).7 This may indicate some genetic heterogeneity of SEDL, although complete skeletal surveys of these six cases were not available for review.
Function of the protein
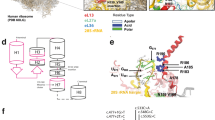
SEDL gene open reading frame of 420 bp encodes a small protein of 140 amino acids. It does not show similarity to any known proteins and there are no conserved protein domains detected. Human recombinant SEDL protein localised to perinuclear membrane structures which partly overlap with VTC (vesicular tubular compartment, known also as ERGIC – ER-to-Golgi intermediate compartment) pointing towards the role of SEDL protein in vesicular transport between ER and the Golgi.6 This function is further supported by studies of the SEDL yeast ortholog, YBR254C (or its protein TRS20). TRS20 is a member of a novel multiprotein complex called transport protein particle (TRAPP) which mediates vesicle docking and fusion.8 Yeast YBR254C knockout experiments show that the yeast SEDL ortholog function is essential for the yeast cell.9 This is not the case for the human SEDL, where patients lacking entire SEDL gene survive (see below). More recently, crystal structure of the human SEDL protein has been determined. This revealed an unexpected similarity to the structures of the N-terminal domain of two SNAREs (soluble N-ethylmaleimide-sensitive factor attachment receptor proteins), Ykt6p and Sec22b, despite no sequence similarity between these proteins and SEDL. A regulatory and/or adaptor role of SEDL protein through multiple protein–protein interactions is proposed.10
Yeast model
There is no animal model for SEDL available however, there is a yeast model. The yeast YBR254C knockout is not viable.9 Preliminary results of complementation experiments with human recombinant SEDL protein show that the human SEDL protein is able to rescue the lethal YBR254C KO phenotype and thus TRS20 protein function (J Gécz, unpublished data).
Mutations
To date 30 different SEDL gene mutations have been identified in 40 unrelated cases. In addition to 21 previously summarised mutations,7 nine have recently been described. These include four new deletions, intron5/exon6del(1371–1445 bp) and intron5/exon6del(750 bp),11 intron2/exon3del12 and 267-271delAAGAC,13 one missense mutation, T248C,14 and four nonsense mutations, G210A and C364T,11 C391T,15 and C329A.16 The most common mutations identified are deletions, accounting for 50% of the types of mutations identified (15/30). The splice site mutation IVS3+5G>A is the most frequently found SEDL mutation to date with five patients identified. SEDL gene mutations are spread along the entire length of the four SEDL coding exons and their flanking introns, that is, exons 3–6. As yet there are no mutations reported in the untranslated exons 1 and 2. There is no obvious correlation between the nature of the mutation and the clinical severity of SEDL. All identified so far mutations SEDL gene appear to have the same effect, loss of function of the SEDL protein, irrespective of their location and type (null mutations, small or large deletions, splice site mutations and missense mutations).
Given the small number of SEDL gene exons and the relative ease of their PCR amplification, direct sequencing is the method of choice for SEDL mutation detection. Primers, which specifically amplify only the X-chromosome gene sequences, have been described.5
Treatment
There is no specific treatment apart from management of the complications of the condition. The most common of these is management of hip dysplasia, which may require hip replacement. Advice should be given regarding prevention of premature arthritis by maintenance of a healthy weight for height and also regular, low-impact exercise such as swimming and cycling. Data regarding the use of the growth hormone in this condition and its effect on final adult height are not available. Significant short stature is usual in this condition and appropriate ongoing psychosocial support of the patient and the family is important. Genetic counselling should be provided to discuss X-linked inheritance. The discovery of the SEDL gene has raised the possibility of carrier testing in females and prenatal testing for this condition.
Electronic database information and accession numbers
(OMIM was accessed at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM,); (XL-SEDT, MIM No. 313400); (SEDL gene, MIM No. 300202, GenBank No. AH008075).
References
Taybi H, Lachman RS : Radiology of syndromes, metabolic disorders, and skeletal dysplasias, 4th ed. St. Louis: Mosby-Year Book Inc., 1996.
Iceton JA, Horne G : Spondylo-epiphyseal dysplasia tarda: the X-linked variety in three brothers. J Bone Joint Surg Br 1986; 68: 616–619.
Hall CM : International Nomenclature Group on Constitutional Disorders of Bone: International nosology and classification of constitutional disorders of bone (2001). Am J Med Genet 2002; 113: 65–77.
Monteiro de Pina Neto J, Bonfim MD, Ferrari I : Classic X-linked spondyloepiphyseal dysplasia tarda in a woman with normal karyotype. Prog Clin Biol Res 1982; 104: 127–132.
Gedeon AK, Colley A, Jamieson R et al: Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat Genet 1999; 22: 400–404.
Gécz J, Hillman MA, Gedeon AK, Cox TC, Baker E, Mulley JC . Gene structure and expression study of the SEDL gene for spondyloepiphyseal dysplasia tarda. Genomics 2000; 69: 242–251.
Gedeon AK, Tiller GE, Le Merrer M et al: The molecular basis of X-linked spondyloepiphyseal dysplasia tarda. Am J Hum Genet 200; 68: 1386–1397.
Sacher M, Jiang Y, Barrowman J et al: TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J 1998; 17: 2494–2503.
Sanjuan R, Leon M, Zueco J, Sentandreu R : Basic phenotypic analysis of six novel yeast genes reveals two essential genes and one which affects the growth rate. Yeast 1999; 15: 351–360.
Jang SB, Kim YG, Cho YS et al: Crystal structure of SEDL and its implications for a genetic disease spondyloepiphyseal dysplasia tarda. J Biol Chem. 2002; 277: 49863–49869.
Christie PT, Curley A, Nesbit MA et al: Mutational analysis in X-linked spondyloepiphyseal dysplasia tarda. J Clin Endocrinol Metab 2001; 86: 3233–3236.
Matsui Y, Yasui N, Ozono K et al: Loss of the SEDL gene product (Sedlin) causes X-linked spondyloepiphyseal dysplasia tarda: identification of a molecular defect in a Japanese family. Am J Med Genet 2001; 99: 328–330.
Mumm S, Christie PT, Finnegan P et al: A five-base pair deletion in the sedlin gene causes spondyloepiphyseal dysplasia tarda in a six-generation Arkansas kindred. J Clin Endocrinol Metab 2000; 85: 3343–3347.
Grunebaum E, Arpaia E, MacKenzie JJ et al: A missense mutation in the SEDL gene results in delayed onset of X linked spondyloepiphyseal dysplasia in a large pedigree. J Med Genet 2001; 38: 409–411.
Takahashi T, Takahashi I, Tsuchida S et al: An SEDL gene mutation in a Japanese kindred of X-linked spondyloepiphyseal dysplasia tarda. Clin Genet 2002; 61: 319–320.
Shi YR, Lee CC, Hsu YA et al: A novel nonsense mutation of the sedlin gene in a family with spondyloepiphyseal dysplasia tarda. Hum Hered 2002; 54: 54–56.
Acknowledgements
We thank G Sutherland and M Shaw for a critical reading of the manuscript. This work was carried out with the support of the National Health and Medical Research Council of Australia.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Savarirayan, R., Thompson, E. & Gécz, J. Spondyloepiphyseal dysplasia tarda (SEDL, MIM #313400). Eur J Hum Genet 11, 639–642 (2003). https://doi.org/10.1038/sj.ejhg.5201025
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201025
Keywords
This article is cited by
-
A novel deletion variant in TRAPPC2 causes spondyloepiphyseal dysplasia tarda in a five-generation Chinese family
BMC Medical Genetics (2020)
-
Novel loss-of-function variants of TRAPPC2 manifesting X-linked spondyloepiphyseal dysplasia tarda: report of two cases
BMC Medical Genetics (2019)
-
Double non-contiguous fractures in a patient with spondylo-epiphyseal dysplasia with spinal ankylosis treated with open and percutaneous spinal fixation technique: a case report
BMC Research Notes (2018)
-
A trapper keeper for TRAPP, its structures and functions
Cellular and Molecular Life Sciences (2012)
-
Noncanonical and canonical splice sites: a novel mutation at the rare noncanonical splice-donor cut site (IVS4+1A>G) of SEDL causes variable splicing isoforms in X-linked spondyloepiphyseal dysplasia tarda
European Journal of Human Genetics (2009)