Abstract
The chromosomal region around marker D8S277 is thought to contribute to susceptibility to hearing impairment in patients with the 1555A>G mutation in mtDNA. We have previously described a family with this mutation, in which some of the members had profound hearing loss, some had a hearing impairment for high-frequency tones and some had completely normal hearing. The phenotypes were thus compatible with a recessive inheritance pattern. We fine-mapped the region around marker D8S277 by sequencing single nucleotide polymorphisms (SNPs) along the 11 Mb region on 8p23, and also sequenced eight defensin genes in the vicinity of D8S277 and the genes GJB2, GJB3, MTO1 and TIMM8A. SNP haplotypes were constructed using the SimWalk2 program. The three persons with a profound hearing loss had identical genotypes in the 11 Mb region on 8p23, but this genotype was also present in a person with normal hearing. The persons with a hearing impairment for high-frequency tones did not share any common haplotype, but one of them shared a genotype with a healthy person. Thus, haplotype comparison excluded a contribution of the region concerned to the expression of hearing impairment in this family, nor could the susceptibility be assigned to the GJB2, GJB3, MTO1 or TIMM8A genes. Extended pedigrees with 1555A>G, such as the present one, provide a good opportunity to identify a modifying nuclear factor. The chromosomal region around 8p23 could be excluded here as the locus for susceptibility to hearing impairment.
Similar content being viewed by others
Introduction
The pharmacological target of aminoglycoside antibiotics is the bacterial ribosome, suggesting that the evolutionarily homologous mitochondrial ribosome is the most likely target of aminoglycoside ototoxicity in the cochlea. Three mutations in the MTRNR1 gene encoding the mitochondrial small ribosomal RNA (12S rRNA) have been found to lead to aminoglycoside-induced maternally inherited sensorineural hearing impairment, the first mutation being 1555A>G.1 Several reports have verified the pathogenic nature of this mutation and have shown that it is the primary cause of the disease and that it has occurred multiple times in human populations. The second mutation, 961delT, together with varying numbers of inserted cytosines, has been reported in a Chinese family2 and in an Italian family3 with aminoglycoside ototoxicity, and a third mutation, 1095T>C, has been found recently.4,5
The homoplasmic 1555A>G mutation is one of the most common causes of nonsyndromic sensorineural hearing loss and aminoglycoside-induced deafness.3 Patients with 1555A>G who receive aminoglycosides have a chance to become deaf,6 but only about 18% of deaf patients with 1555A>G have been treated with aminoglycosides.6 In the absence of aminoglycosides, the mutation leads to a phenotype that ranges from severe congenital deafness to completely normal hearing.1,7 Thus, expression of the deafness phenotype in patients with 1555A>G appears to require contributions from environmental factors or nuclear modifier gene(s).1,7,8,9,10
The nuclear background may determine the expression of the disease, but genetic susceptibility may be heterogeneous and have a complex model of inheritance.11 Several genes have been associated with hearing impairment. Mutations in the connexin 26 gene (GJB2) have been assumed to aggravate deafness in Japanese patients with 1555A>G,12 although the role of this gene has been refuted in other studies.11,13 Termination and substitution mutations in this gene have been identified in autosomal dominant and autosomal recessive nonsyndromic deafness.14,15,16 A promising candidate gene has recently been identified, with the observation that mutant alleles of the yeast MTO1 gene, encoding a mitochondrial protein, manifest a respiratory-deficient phenotype only when coupled with the mitochondrial 15S rRNA PR454 mutation, corresponding to the human 1555A>G mutation.17
A genomewide linkage search has suggested a modifier locus on chromosome 8 in families with 1555A>G.18 Nonparametric analysis supported the role of the chromosomal region around marker D8S277, but there are no obvious candidate genes in this locus, and the putative modifier nuclear gene remains to be identified. We report here on studies of the locus on chromosome 8 in a large family with the homoplasmic 1555A>G mutation, the audiometric pattern of which is compatible with the hypothesis that heterozygotes have a hearing impairment for high-frequency tones, while homozygotes have profound hearing loss. We fine-mapped the locus by sequencing single nucleotide polymorphisms (SNPs) along an 11 Mb region on 8p23 and by sequencing the exons of eight defensin genes in the vicinity of D8S277. In addition, we sequenced other candidate genes elsewhere in the genome, including GJB2 and MTO1. Since all three family members with profound hearing loss were men, we also sequenced an X-chromosomal gene, a homologue to yeast translocase of inner mitochondrial membrane 8A (TIMM8A). A mutation is found in this gene in Deafness Dystonia Syndrome (MTS/DFN-1), which has been postulated to be caused by a defective mitochondrial protein import system.19,20,21 Finally, we also sequenced connexin 31 (GJB3) on chromosome 1, since mutations in this gene cause deafness characterized by progressive high-frequency hearing loss in adulthood and milder expression in females.22
Methods
The pedigree
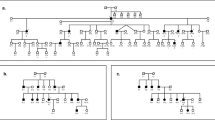
Audiometry was performed on members of a family with the homoplasmic 1555A>G mtDNA mutation (Figure 1).23 Three sons in the third generation had profound hearing loss with onset at the age of 4–5 years. Four of their siblings had a hearing impairment for high-frequency tones with onset in youth or early adulthood, while five of the siblings had normal hearing. The mother and two of her siblings also had hearing impairment for high-frequency tones. There was no evidence of the use of aminoglycoside antibiotics in this family. DNA samples from three family members had been included in a previous linkage study,24 where an autosomal modifier locus was detected on chromosome 8. The family was potentially linked to the region with the NPL>0.8. The patients were unaffected neurologically.
Pedigree of the family with 1555A>G and SNP haplotypes constructed for 8p23. First line below the pedigree symbols, subject identification; second line, age at audiometry examination. Solid symbols, profound hearing impairment; half-filled symbols, hearing impairment at high frequencies; open symbols, normal hearing. Eight haplotypes were characterized in this family. The inset shows the letter codes used for them. One recombination was discovered in the maternal allele of III/10. Patients III/4, III/6 and III/8 had been included in the linkage analysis reported previously.24
Molecular methods
Sequenced SNPs and candidate genes
A nonparametric linkage analysis has supported the notion of susceptibility locus in the chromosomal region around marker D8S277 in patients with 1555A>G and hearing impairment.24 We fine-mapped this locus by sequencing SNPs along the 11 Mb region on 8p23 (Figure 2). Seven regions with multiple known SNPs were selected from the NCBI database (http://www.ncbi.nlm.nih.gov, GenBank accession numbers; nucleotides in parentheses): (a) (AC005010; 131235–131728), (b) (AF215849; 130555–131146), (c) (AC016892; 118792–119253), (d) (AF200455; 24604–24934), (e) (AC130346; 65205–65424), (f) (AC016485; 57857–58384), (g) (AC010400; 33756–34076). Furthermore, exons and exon–intron boundaries of eight defensin genes in the vicinity of D8S277 were sequenced (DEFA1, GenBank accession number L12690; DEFA3, L12691; DEFA4, U18745; DEFA5, M97925; DEFA6, U33317; DEFB1, U50930; DEFB3, AF252830; and DEFB4, AF040153).
Physical map of the chromosomal region 8p23. The physical locations of the selected genes and hypothetical genes are shown in relation to the sequenced SNP regions (a – g) and to microsatellite markers. The DEFA cluster denotes defensin alpha genes in the order DEFA6, DEFA4, DEFA1, DEFA3 and DEFA5, and the DEFB cluster denotes the defensin beta genes DEFB3 and DEFB4. Nonparametric linkage analysis supports a role for the chromosomal region around marker D8S277 in 1555A>G-associated hearing impairment susceptibility. The map was generated according to the NCBI MapViewer, build 30, based on sequence information available on 24 June 2002 (http://www.ncbi.nlm.nih.gov).
In addition to the locus on chromosome 8, we sequenced the putative nuclear modifier gene MTO1 (GenBank accession number; AF319422) on chromosome 6, two connexin genes: GJB2 (connexin 26, GenBank accession number; M86849) on chromosome 13q11 and GJB3 (connexin 31, GenBank accession number; AF052692) on chromosome 1p34, and TIMM8A (GenBank accession number; U66035) on chromosome Xq22.
PCR and sequencing
SNPs and candidate genes were sequenced in nine affected persons, three of whom had profound hearing loss, and in seven unaffected persons. Total DNA was extracted from the blood cells using Phase Lock Gel (Eppendorf AG, Hamburg, Germany) with phenol and chloroform.
Each fragment with SNPs was amplified from genomic DNA by PCR using a pair of specific primers (Table 1). The reactions were performed in a total volume of 50 μl, containing 40 ng of template DNA, 1 × AmpliTaq Gold DNA polymerase buffer (Applied Biosystems, Foster City, CA, USA), 0.2 mM of each nucleotide, 1 μM of primers, 2.5 mM of MgCl2, 0.9 M dimethylsulfoxide (DMSO) and 1 U AmpliTaq Gold DNA polymerase. PCR was performed with initial denaturation for 15 min at 95°C followed by 38 cycles of denaturation, annealing and extension. Each cycle consisted of 40 s denaturation at 94°C, annealing for 40 s at temperatures ranging from 63°C to 55°C with a 0.5°C reduction per cycle, followed by 21 cycles at 55°C, and extension for 45 s at 72°C. There was a final extension at 72°C for 10 min.
In order to verify the changes in sequences, especially the heterozygous ones, the sequencing was performed in both directions. After treatment with exonuclease I (New England Biolabs, Beverly, MA, USA) and shrimp alkaline phosphatase (Amersham Pharmacia Biotech, Piscataway, NJ, USA) the amplified DNA fragments were sequenced using a DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Pharmacia Biotech) and analyzed by automated sequencing (ABI PRISM™ 377 Sequencer, Applied Biosystems). The sequence comparisons were performed by Sequencher 4.0.5 (Gene Codes Corporation, Ann Arbor, MI, USA).
Haplotype construction
We compiled the SNP data from sequenced SNP-rich regions and those found in eight defensin genes (Table 2). The SNP haplotypes were constructed using the SimWalk2 v. 2.82 program.25 We monitored haplotype sharing in persons with profound hearing loss or hearing impairment for high-frequency tones by comparison with persons having normal hearing. The haplotypes of GJB3 gene were constructed in a similar fashion. The computations for the analyses were performed using programs available at CSC – The Finnish Information Technology Center for Science (http://www.csc.fi).
Ethical considerations
All of the participants had received information on their disease, and blood samples were collected after obtaining informed consent. Family, members below the age of 18 years were excluded. The protocols were approved by the Ethics Committee of the Medical Faculty of University of Oulu.
Results
Fine–mapping of 8p23
A non parametric linkage analysis has supported the notion of a role for the chromosomal region around marker D8S277 in 1555A>G-associated hearing impairment susceptibility.24 We have previously described a family with homoplasmic 1555A>G that is compatible with the hypothesis that heterozygotes have a hearing impairment for high-frequency tones, while homozygotes have profound hearing loss. We fine-mapped this locus here by sequencing SNPs along the 11 Mb region on 8p23. Furthermore, we sequenced exons and exon–intron boundaries of eight defensin genes in the vicinity of D8S277. The compiled SNP data from sequenced SNP-rich regions and those found in the eight defensin genes were used to construct SNP haplotypes using the SimWalk2 program (Figure 1, Table 2). All the siblings in the second generation share the common allele a from the mother, and all the affected persons have an allele combination a/c, with the c allele from the father. In the third generation, the c allele from the grandfather is found in both the affected and unaffected persons. All three persons with profound hearing loss share the same allele combination a/f, which is also found in III/1, with completely normal hearing at the age of 35 years. Furthermore, a healthy sibling (III/4) and a sibling (III/5) with a hearing impairment for high-frequency tones shared the genotype a/e. Thus, haplotype comparison enabled us to exclude a contribution of the region concerned to the expression of 1555A>G-associated hearing impairment in this family.
We found a total of 12 single nucleotide changes in the eight defensin genes (Table 2). Nine of them were known polymorphisms and two were synonymous polymorphisms that have not been reported previously. A transition in exon 1 of the defensin B4 gene led to the replacement of the second methionine by isoleucine. This polymorphism was found in heterozygous form in II/1, II/2, II/4, III/3, III/7 and III/9 and in homozygous form in III/10. All three phenotypes were present in these subjects.
Sequencing of candidate genes in other genes
Eight variable positions were detected in the GJB3 gene. Haplotype analysis of its SNPs excluded this gene and the region 1p34 (data not shown). There were no changes in the coding exon of GJB2, and the only change in its vicinity was an A>G transition in the 3′ UTR, which was homozygous in all persons except for III/10. The only change in MTO1 compared with the reference sequence was an A>G transition in the intron between exons 5 and 6, which was homozygous in all family members. We could not find any changes in the exons or exon flanking regions of TIMM8A.
Discussion
We could not verify the association of the locus D8S277 with hearing impairment caused by the mitochondrial 1555A>G mutation in this large Finnish pedigree. The family has been potentially linked to the region with the NPL>0.824 in a linkage study which included members of the pedigree. Haplotype analysis of 8p23 revealed, however, that the two affected persons shared both alleles, while the unaffected person shared only one allele with them (Figure 1). These three samples were thus compatible with the results of the linkage study, but samples from other family members disproved the association, assuming that the hypothesis of a recessive inheritance pattern is correct and the penetrance of the phenotype is reasonable. Genetic segregation analysis of the Arab–Israeli pedigree has indicated the presence of an autosomal recessive factor responsible for the phenotypic expression.26 The phenotype extended from severe, profound congenital hearing loss to mild and/or late-onset hearing loss and normal hearing.26,27 Even though the penetrance of the nuclear modifying factor may not be complete, it is unlikely that one of the siblings with entirely normal hearing (III/1) could have the same alleles as persons with childhood-onset profound hearing loss, and a healthy sibling (III/4) could have the same alleles with a sibling (III/5) with a hearing impairment for high-frequency tones.
Two genomewide linkage analyses of families with 1555A>G have revealed several putative loci, but none of them were shared between the studies.11,18 Four loci (D1S234, D13S221, D4S1627, D17S836) showed slightly suggestive LOD scores in a large Arab–Israeli family,11 and the second genomewide linkage screening study identified two loci (D8S277, D9S175) with two-point LOD scores of 1.5 and 1.2 and multipoint LOD scores of 1.3 and 1.8 in the autosomal recessive model.18 Inclusion of new families increased the nonparametric multipoint LOD score of D8S277 to 4.1.24 Mitochondrial ribosomal proteins would be good candidates for a modifier, but none of these genes have been mapped to chromosome 8.28 However, the nuclear modifier gene may as well code for a protein involved in a cochlea-specific, energy-requiring process. Thus, mild defect of mitochondrial translation and a mild defect in a nuclear-encoded function could together lead to hearing loss.
Our results refuted the role of two connexin genes, GJB2 and GJB3, a promising new candidate gene MTO117 and the TIMM8A gene as contributors to the hearing impairment in the pedigree with 1555A>G. The pedigree data could be compatible with an X-chromosomal factor, since all three family members with profound hearing loss were men. Sequencing of TIMM8A, an X-chromosomal gene in which a mutation is found in Deafness Dystonia Syndrome,19,20,21 did not reveal any variants. Interestingly, mutations in connexin 31 (GJB3) on chromosome 1 cause deafness characterized by progressive high-frequency hearing loss in adulthood and milder expression in females.22
Furthermore, the GJB3 gene is located on 1p34, together with D1S234, which gave the best results in the two-point and multipoint analyses in the autosomal recessive model for the Arab–Israeli family.11 The haplotype analysis thus enables us to refute this region. Neither GJB2 nor MTO1 contributed to the penetrance of the deafness phenotype in the Finnish family.
The phenotypic and genotypic features of Leber's hereditary optic neuroretinopathy (LHON) resemble those of the hearing impairment associated with 1555A>G. Both diseases affect primarily one organ in the sensory system and both are caused by mtDNA mutations that occur mainly in the homoplasmic state. Interestingly, there is an excess of affected men in LHON pedigrees. This observation has suggested an X-linked visual loss susceptibility gene, a hypothesis that was supported by linkage studies in Finnish pedigrees,29 but subsequent linkage studies have excluded the presence of such a locus.30,31 The resemblance between LHON and the hearing impairment associated with 1555A>G may suggest that these two mitochondrial conditions share a common susceptibility factor.
It is likely that the interaction of genes with environmental factors plays an important role in the development of deafness, especially in late-onset cases.6 The only environmental factor determined to date is the administration of aminoglycoside antibiotics, which accounts for about 18% of deafness in patients with 1555A>G.6 Furthermore, deafness shows an age-dependent penetrance. The probability of deafness at 30 years of age is 97% among those who have received aminoglycosides and 40% among those who have not.6 Late-onset deafness suggests the involvement of other environmental factors in genetically susceptible individuals. Such a trigger could be a viral infection, for example, exposure to chemicals other than aminoglycosides, or exposure to noise – factors that are more common in the environment than aminoglycosides. Extended pedigrees, such as the Finnish pedigree studied here or the Arab–Israeli pedigree,26 are advantageous when studying the genetics of such a disease and may give more accurate results in attempts to identify a modifying nuclear factor, because the genetic heterogeneity is low and the environmental factors are alike.
References
Prezant TR, Agapian JV, Bohlman MC et al: Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 1993; 4: 289–294.
Bacino C, Prezant TR, Bu X, Fournier P, Fischel-Ghodsian N : Susceptibility mutations in the mitochondrial small ribosomal RNA gene in aminoglycoside induced deafness. Pharmacogenetics 1995; 5: 165–172.
Fischel-Ghodsian N : Genetic factors in aminoglycoside toxicity. Ann N Y Acad Sci 1999; 884: 99–109.
Thyagarajan D, Bressman S, Bruno C et al: A novel mitochondrial 12SrRNA point mutation in parkinsonism, deafness, and neuropathy. Ann Neurol 2000; 48: 730–736.
Tessa A, Giannotti A, Tieri L, Vilarinho L, Marotta G, Santorelli FM : Maternally inherited deafness associated with a T1095C mutation in the mDNA. Eur J Hum Genet 2001; 9: 147–149.
Estivill X, Govea N, Barcelo E et al: Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet 1998; 62: 27–35.
Fischel-Ghodsian N, Prezant TR, Bu X, Oztas S : Mitochondrial ribosomal RNA gene mutation in a patient with sporadic aminoglycoside ototoxicity. Am J Otolaryngol 1993; 14: 399–403.
Hutchin T, Haworth I, Higashi K et al: A molecular basis for human hypersensitivity to aminoglycoside antibiotics. Nucleic Acids Res 1993; 21: 4174–4179.
Guan MX, Fischel-Ghodsian N, Attardi G : Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet 1996; 5: 963–971.
Guan MX, Fischel-Ghodsian N, Attardi G : Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet 2001; 10: 573–580.
Bykhovskaya Y, Shohat M, Ehrenman K et al: Evidence for complex nuclear inheritance in a pedigree with nonsyndromic deafness due to a homoplasmic mitochondrial mutation. Am J Med Genet 1998; 77: 421–426.
Abe S, Kelley PM, Kimberling WJ, Usami S : Connexin 26 gene (GJB2) mutation modulates the severity of hearing loss associated with the 1555A>G mitochondrial mutation. Am J Med Genet 2001; 103: 334–338.
Lopez-Bigas N, Rabionet R, Martinez E et al: Mutations in the mitochondrial tRNA Ser(UCN) and in the GJB2 (connexin 26) gene are not modifiers of the age at onset or severity of hearing loss in Spanish patients with the 12S rRNA A1555G mutation. Am J Hum Genet 2000; 66: 1465–1467.
Kelsell DP, Dunlop J, Stevens HP et al: Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 1997; 387: 80–83.
Zelante L, Gasparini P, Estivill X et al: Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 1997; 6: 1605–1609.
Denoyelle F, Lina-Granade G, Plauchu H et al: Connexin 26 gene linked to a dominant deafness. Nature 1998; 393: 319–320.
Li X, Li R, Lin X, Guan MX : Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12S rRNA A1555G mutation. J Biol Chem 2002; 277: 27256–27264.
Bykhovskaya Y, Estivill X, Taylor K et al: Candidate locus for a nuclear modifier gene for maternally inherited deafness. Am J Hum Genet 2000; 66: 1905–1910.
Jin H, May M, Tranebjaerg L et al: A novel X-linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nat Genet 1996; 14: 177–180.
Hofmann S, Rothbauer U, Muhlenbein N et al: The C66W mutation in the deafness dystonia peptide 1 (DDP1) affects the formation of functional DDP1–TIM13 complexes in the mitochondrial intermembrane space. J Biol Chem 2002; 277: 23287–23293.
Roesch K, Curran SP, Tranebjaerg L, Koehler CM : Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum Mol Genet 2002; 11: 477–486.
Xia J, Liu C, Tang B et al: Mutations in the gene encoding gap junction protein beta-3 associated with autosomal dominant hearing impairment. Nat Genet 1998; 20: 370–373.
Lehtonen M, Uimonen S, Hassinen IE, Majamaa K : Frequency of mitochondrial DNA point mutations among patients with familial sensorineural hearing impairment. Eur J Hum Genet 2000; 8: 315–318.
Bykhovskaya Y, Yang H, Taylor K et al: Modifier locus for mitochondrial DNA disease: linkage and linkage disequilibrium mapping of a nuclear modifier gene for maternally inherited deafness. Genet Med 2001; 3: 177–180.
Sobel E, Lange K : Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 1996; 58: 1323–1337.
Jaber L, Shohat M, Bu X et al: Sensorineural deafness inherited as a tissue specific mitochondrial disorder. J Med Genet 1992; 29: 86–90.
Braverman I, Jaber L, Levi H et al: Audiovestibular findings in patients with deafness caused by a mitochondrial susceptibility mutation and precipitated by an inherited nuclear mutation or aminoglycosides. Arch Otolaryngol Head Neck Surg 1996; 122: 1001–1004.
O'Brien TW, Liu J, Sylvester JE et al: Mammalian mitochondrial ribosomal proteins (4). Amino acid sequencing, characterization, and identification of corresponding gene sequences. J Biol Chem 2000; 275: 18153–18159.
Vilkki J, Ott J, Savontaus ML, Aula P, Nikoskelainen EK : Optic atrophy in Leber hereditary optic neuroretinopathy is probably determined by an X-chromosomal gene closely linked to DXS7. Am J Hum Genet 1991; 48: 486–491.
Carvalho MR, Muller B, Rotzer E et al: Leber's hereditary optic neuroretinopathy and the X-chromosomal susceptibility factor: no linkage to DXS7. Hum Hered 1992; 42: 316–320.
Sweeney MG, Davis MB, Lashwood A, Brockington M, Toscano A, Harding AE : Evidence against an X-linked locus close to DXS7 determining visual loss susceptibility in British and Italian families with Leber hereditary optic neuropathy. Am J Hum Genet 1992; 51: 741–748.
Acknowledgements
This work was supported partly by grants from the Medical Research Council of the Academy of Finland, the Sigrid Juselius Foundation and the Multiple Sclerosis Foundation of Finland. The expert technical assistance of Mrs. Irma Vuoti, Mrs. Pirjo Keränen and Mrs. Anja Heikkinen is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Finnilä, S., Majamaa, K. Lack of a modulative factor in locus 8p23 in a Finnish family with nonsyndromic sensorineural hearing loss associated with the 1555A>G mitochondrial DNA mutation. Eur J Hum Genet 11, 652–658 (2003). https://doi.org/10.1038/sj.ejhg.5201017
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201017