Abstract
Juvenile hemochromatosis (JH) is a rare autosomal recessive disorder that causes iron overload. In the French Canadian region of Saguenay Lac-Saint-Jean the worldwide largest cohort of JH cases has been identified. Here, we report the mapping of this large cohort of cases to the HFE2 locus on chromosome 1q. A maximum multipoint location score of 7.02 was observed with marker D1S2344. A common ancestral haplotype, showing the presence of a founder effect, was identified. The analysis of recombinants allowed us to confirm the JH candidate region.
Similar content being viewed by others
Introduction
Juvenile hemochromatosis, or hemochromatosis type II (JH; MIM 602390), is an inborn error of iron metabolism, which leads to severe iron loading and organ failure at age before 30 years. First symptoms include heart failure, cardiac arrhythmia and hypogonadotropic hypogonadism.1,2,3 JH is a rare disorder, characterized by an autosomal recessive inheritance, distinct from classic-type hemochromatosis or HFE (MIM 235200).4,5
JH patients do not show linkage to chromosome 6p and do not have mutations in the HFE gene that lead to the common hereditary hemochromatosis.1,6,7 Although the gene associated with this disorder has not been identified, the locus maps to chromosome 1q, in a 4 cM interval between the markers D1S442 and D1S2347.8,9
Further studies showed the presence of genetic heterogeneity in other population specific groups of patients.10,11
Very recently, two mutations in the antimicrobial peptide Hamp gene, which maps to chromosome 19, have been found in affected members from two JH families. This result increases the genetic heterogeneity of iron-overload diseases and, moreover, identifies a new form of juvenile hemochromatosis.12
At the beginning of the 1990s, we conducted a clinical, biochemical and genetic study on 30 families with hereditary hemochromatosis in the Saguenay Lac-Saint-Jean region of Quebec.13,14,15,16 In 2000, 32 probands were genotyped for mutations in the HFE gene. A high proportion (28.1%) of hemochromatosis chromosomes carried no known or new mutations.17,18 In fact, most of these patients fulfilled the JH criteria (17 patients distributed in 12 families).
Here, we report a linkage analysis study performed on a cohort of JH patients from the Saguenay Lac-Saint-Jean region. Data obtained allowed us to confirm linkage to the JH locus on chromosome 1q.
Materials and methods
JH patients
A total of 17 JH patients (seven females and 10 males) belonging to 12 families were included in the study (Figure 1). The average age at diagnosis was 24.1 years. The diagnosis was made on the basis of clinical history, physical examination and persistently raised iron indices (% transferrin saturation and serum ferritin). Liver biopsy was performed in at least one affected patient of each sibship. The clinical, biochemical, and genetic characteristics were as previously described.1
Molecular studies
EDTA blood samples of 97 individuals were collected for genetic studies after informed consent was obtained. DNA was extracted according to standard protocols. Classical PCR reactions were carried out on 100 ng DNA. PCR fragments were analysed on ABI PRISM 3100 and results processed by GENESCAN and GENOTYPER version 2.5 softwares from ABI PRISM. According to available maps (GDB and CELERA databases), we selected 13 microsatellites of chromosome 1q. The following polymorphic markers were used: D1S2875, D1S2863, D1S514, D1S185, D1S2696, D1S442, D1S2344, D1S1156, D1S2222, GATA13C08, D1S498, D1S2347 and D1S2343.
Linkage analysis
Statistical analysis was carried out on the assumption of autosomal recessive disorder with complete penetrance. The disease-gene frequency was set to 0.001, and all marker alleles were considered to be equally frequent. Two-point linkage analyses were performed with ILINK and MLINK programs, version 5.1, from the LINKAGE software package.19,20 The input data files for the multipoint analysis were created using MEGA 2 program.21 The multipoint and haplotype analyses were performed using SIMWALK 2 v.2.82 that calculates the Maximum Location Score (directly comparable with multipoint LOD score) using simulated annealing and MCMC (Monte Carlo Markov Chain) algorithm.22 We assigned a specific allele number for each marker within each family to reduce the time for the analysis.
Results
Linkage analysis of JH locus
After a linkage study using microsatellite markers from the HFE2 show significant pairwise lod scores in our French Canadian cohort have been detected, as shown in Table 1. In particular, a maximum LOD score of 4.07 at θmax=0.0 was obtained with markers D1S2344 and D1S1156 (Table 1). Multipoint linkage analysis performed with markers D1S514, D1S185, D1S2696, D1S442, D1S2344, D1S1156, GATA13C08, D1S498 and D1S2347 showed a maximum location score of 7.02 with markers D1S2344 and D1S1156.
Recombinantional events and haplotype analysis
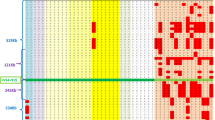
In order to restrict the HFE2 candidate region we tried to place additional microsatellites and SNP markers in the region defined by D1S442 and GATA13C08 markers. Only marker D1S2222 located within this interval but, unfortunately, it was not informative with Zmax=0.00 at θmax=0.503 (Table 1). A common haplotype in all patients was obtained as shown in Figure 2. A recombinantional event was found in individual 2020 in family 27 with markers D1S514, D1S185, D1S2696, D1S442, D1S2344 and D1S1156. A heterogeneous genotype for D1S442 was found in patient 2014 of the same family and another one in patient 2065 of family 48 with the markers D1S2875, D1S2863, D1S514, D1S185, D1S2696 and D1S442 (Figure 2). These recombinational events confirm both the centromeric limit of JH region at marker D1S442 and the telomeric one at marker GATA13CO8. The same telomeric limit was observed in family 58 (data not shown). This critical region spans within a 4 cM interval between markers D1S442 and GATA13CO8.
Discussion
Several hereditary disorders have been shown to have a high frequency in the French Canadian population of Saguenay Lac-Saint-Jean, a geographically isolated region located 200 km north-east of Quebec City and surrounded by tracts of unsettled land and forest.13,16 The values of the mean inbreeding and kinship coefficients were much higher among patients with hemochromatosis than those with other autosomal recessive disorders in Saguenay Lac-Saint-Jean.16 Therefore, combined with the clinical picture, it became very quickly evident, that a different locus than HFE1 was involved in this population. Here, we demonstrate that a large cohort of patients fulfilling the diagnostic criteria of Juvenile Hemochromatosis and coming from Saguenay Lac-Saint-Jean region confirm and better define the HFE2 locus on chromosome 1. In addition, we were able to demonstrate the presence of a common ancestral haplotype which should be the consequence of a strong founder effect. This finding clearly suggests the presence of a unique mutation in our Saguenay Lac-Saint-Jean population with JH.
Unfortunately, no candidate gene is included in the HFE2 region. Nevertheless, 10 genes have been so far selected and analyzed by DHPLC. Negative data have been obtained in all cases. Additional work is now in progress to select more genes using both standard and new approaches, such as microarrays.
In conclusion, the present data demonstrate that the largest worldwide cohort of JH families maps to HFE2. This cohort is the result of a strong founder effect and is useful to further refine the HFE2 candidate region and we confirm the juvenile hemochromatosis locus maps on chromosome 1q.
References
Rivard SR, Mura C, Simard H et al: Clinical and molecular aspects of Juvenile hemochromatosis in Saguenay-Lac-Saint-Jean (Québec, Canada): Blood Cells, Mol Dis 2000; 26: 10–14.
Camaschella C: Juvenile hemochromatosis. Baillieres Clin Gastroenterol 1998; 12: 227–235.
Cazzola M, Ascari E, Barosi G et al: Juvenile idiopathic hemochromatosis: a life-threatening disorder presenting as hypogonadotropic hypogonadism. Hum Genet 1983; 65: 149–154.
Camaschella C, Roetto A, Cicilano M et al: Juvenile and adult hemochromatosis are distinct genetic disorders. Eur J Hum Genet 1997; 5: 371–375.
De Gobbi M, Roetto A, Piperno A et al: Natural history of Juvenile Hemochromatosis. Br J aematol 2002; 117(4): 973–979 (Review).
Carella M, D’Ambrosio L, Totaro A et al:. Mutation analysis of HLA-H gene in Italian hemochromatosis patients. Am J Hum Genet 1997; 60: 828–832.
Ferder JN, Gnirke A, Thomas W et al: A novel MHC class I-like gene is mutated in patients with hereditary hemochromatosis. Nat Genet 1996; 13: 399–408.
Roetto A, Totaro A, Cazzola M et al: Juvenile hemochromatosis locus maps to chromosome1q. Am J Hum Genet 1999; 64: 1388–1393.
Papanikolaou G, Politou M, Roetto A et al: Linkage to chromosome 1q in Greek families with juvenile hemochromatosis. Blood Cells Mol Dis 2001; 27: 744–749.
Papanikolaou G, Papaioannou M, Politou M et al: Genetic heterogeneity underlies juvenile hemochromatosis phenotype: analysis of three families of northern greek origin. Blood Cells Mol Dis 2002; 29: 168–173.
Barton JC, Rao SV, Pereira NM et al: Juvenile hemochromatosis in the southeastern United States: a report of seven cases in two kinships. Blood Cells Mol Dis 2002; 29: 104–115.
Roetto A, Papanikolaou G, Politou M et al: Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet 2003; 33: 21–22.
De Braekeleer M, St-Pierre C, Vigneault A et al: Hemochromatosis and pyruvate kinase deficiency. Report of a case and literature review. Ann Hematol 1991; 62: 188–189.
De Braekeleer M, Vigneault A, Roy R, Simard H: Hereditary hemochromatosis and HLA antigens in Saguenay-Lac-Saint-Jean (Québec, Canada). 1992; Repport, Laboratoire de recherches en Épidémiologie Génétique, Université du Québec à Chicoutimi.
De Braekeleer M: A prevalence and fertility study of hemochromatosis in Saguenay-Lac-Saint-Jean. Ann Hum Biol 1993; 20: 501–505.
Braekeleer M: Inbreeding, kinship and surnames in hereditary disorders: the experience in Saguenay-Lac-Saint-Jean (Québec). Collegium Antropologicum 1995; 19: 289–304.
Rivard SR, Mura C, Simard H et al: Mutation analysis in the HFE gene in patients with hereditary hemochromatosis in Saguenay-Lac-Saint-Jean (Québec, Canada). Br J Haematol 2000; 108: 854–858.
Lathrop GM, Lalouel JM, Julier C, Ott J : Related articles, links strategies for mulitilocus linkage analysis in humans. Proc Natl Acad Sci USA 1984; 81: 3443–3446.
Lathrop GM, Lalouel JM: Related articles, Links easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 1984; 36: 460–465.
Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP,, Weeks DE: Mega2, a data-handling program for facilitating genetic linkage and association analyses. Am J Hum Genet 1999; 65: A436.
Sobel E, Lange K: (SIMWALK2) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 1996; 58: 1323–1337.
De Braekeleer M . Autosomal recessive disorders in Saguenay-Lac-Saint-Jean, Québec: A study of inbreeding. Ann Hum Genet 1996; 60(PC1): 51–56.
Acknowledgements
This research was partially granted by EEC FP5- PISDAP QLTR-1999-02237 grant (to PG). RF is a recipient of a Second University of Naples research contract on this topic, and SAGEN Pharma in Canada. The authors thank Mrs Julie Lavoie, Maryse Beaudet and Angela D’Eustacchio for their invaluable help.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rivard, S., Lanzara, C., Grimard, D. et al. Juvenile hemochromatosis locus maps to chromosome 1q in a French Canadian population. Eur J Hum Genet 11, 585–589 (2003). https://doi.org/10.1038/sj.ejhg.5201009
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201009
Keywords
This article is cited by
-
Design of high-performance parallelized gene predictors in MATLAB
BMC Research Notes (2012)
-
Two-marker association tests yield new disease associations for coronary artery disease and hypertension
Human Genetics (2011)
-
Genome-wide searching of rare genetic variants in WTCCC data
Human Genetics (2010)
-
Molecular Evolution of Hemojuvelin and the Repulsive Guidance Molecule Family
Journal of Molecular Evolution (2007)
-
Hemojuvelin: a supposed role in iron metabolism one year after its discovery
Journal of Molecular Medicine (2005)