Abstract
In vitro folliculogenesis of cryopreserved ovarian tissue could be an effective method for insuring fertility for patients who receive gonadotoxic treatment. Although several culture systems have been described for growing female gametes in vitro, the production of competent oocytes for further development remains a considerable challenge. The purpose of our study was to determine whether maternal primary imprinting progresses normally during mouse oocyte growth in vitro. We analysed the DNA methylation status of differentially methylated regions of the imprinted genes H19, Mest/Peg1 and Igf2R using fully grown germinal vesicle-stage oocytes (fg oocytes) produced by in vitro folliculogenesis from early preantral follicles. When compared to fg oocytes removal from control females, we observed after in vitro development, a loss of methylation at the Igf2R locus in six out of seven independent experiments and Mest/Peg1 locus (one out of seven), and a gain of methylation at the H19 locus (one out of seven). These results provide insight into the dysregulation of the process of primary imprinting during oocyte growth in vitro and highlight the need for effective new biomarkers to identify complete nuclear reprogramming competence after in vitro folliculogenesis.
Similar content being viewed by others
Introduction
Various culture systems have been described for the maturation of female gametes in vitro.1 However, the development of primordial follicles to maturity, in which oocytes acquire in vitro competence to undergo maturation, fertilization and embryonic development, remains a considerable challenge. Recent evidence suggests that mechanisms regulating oocyte growth underlie the establishment of maternal primary imprinting during mouse oogenesis.2 This lead to monoallelic expression/repression of imprinted genes involved in embryonic growth, placentation and behavioral development.3 During oogenesis, expression and/or methylation analysis of imprinted genes provide evidence for the erasure of the parental imprint in primordial germs cells.4 New imprints are then later initiated during the development of nongrowing oocytes to fully grown (fg) oocytes as indicated by a marked change of maternal germline-specific imprints.2,5 Thus, maternal primary imprinting may be particularly susceptible to methylation changes that occur at imprinted loci during oogenesis. Since previous studies in mice have suggested that culture conditions could affect the epigenetic regulation of genomic imprinting,6,7 the aim of our study was to determine the consequence of in vitro folliculogenesis on the progress of maternal primary imprinting during germinal vesicle-stage oocyte growth.
Materials and methods
Preantral follicles containing early growing germinal vesicle-stage oocytes (eg oocytes) were isolated from ovaries of 11- and 12-day old F1 hybrid female mice (C57bl6 × CBA). Early preantral follicles were selected according to the following criteria: intact follicle structure with one or two layers of granulosa cells, visible round and central oocyte, adhering thecal cells, follicle diameter between 100 and 130 μm. Follicles were placed individually into 10 μl drops of culture medium, under pre-equilibrated mineral oil (Sigma-Aldrich) at 37°C in an atmosphere of 5% CO2 in air, using a system similar to the one previously described.8 Culture medium consisted of α minimal essential medium (Life Technologies) supplemented with 5% heat inactivated FCS, 10 μg/ml transferrin (Boehringer Mannheim, France), 5 μg/ml insulin (Boehringer Mannheim) and 100 mIU/ml r-FSH (Serono, France). Healthy oocyte–cumulus complexes consisting of compact granulosa cells with regular borders surrounding the entire mass were used in these experiments. On day 11 of culture, fg oocytes were freed of their surrounding cumulus cells by repeated pipetting in 1 mg/ml hyaluronidase (Sigma-Aldrich) and washed in serum-free modified HTF medium (Irvine Scientific, USA). As control, we used antral follicle grown in vivo collected on the surface of the ovary of adult mice.
Bisulfite treatment of 5–8 fg oocytes imbedded in agarose beads was performed as previously reported.9 Special care was taken to avoid contamination from granulosa cells. DNA methylation analysis of specific differentially methylated regions (DMRs) of H19, Mest/Peg1 and Igf2R was performed on independent pools of 5–8 fg oocytes by denaturing high-performance liquid chromatography (DHPLC) analysis as recently reported.10
Results and discussion
We have analysed the methylation status of DMRs of imprinted genes using a newly developed DHPLC-based procedure.10 Analysis of control fg oocytes from different PCR products indicated that H19 alleles remained essentially all unmethylated (Figure 1c), whereas Mest/Peg1 alleles (Figure 2c) and Igf2R alleles (Figure 3c) were almost all methylated when compared to curves representing 100 and 0% methylated/unmethylated control alleles. These methylation profiles were characteristic of the maternal imprint and provided evidence that the maternal methylation imprint was already established in in vivo fg oocytes as previously described.2,5,11
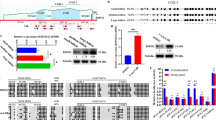
Methylation analysis of imprinted H19, (a) Genomic structures of the DMR studied here. We amplified 13 CpGs of the H19 imprinting control region (accession no. U19619.1; nt −2975, −3204), Arrow, transcription start site of the gene; black box, exons; small black squares, individual CpGs within the areas amplified. (b) DHPLC chromatograms of PCR products of control samples. Methylation profiles were studied by newly developed DHPLC-based method, as recently described.10 Single peaks consisting of homoduplexes characterized homogeneous samples with methylated or unmethylated alleles, whereas multiple peaks consisting of homoduplex and heteroduplex formations characterized heterogeneous samples: (*), heteroduplexes; granulosa cells; control methylated alleles; control unmethylated alleles. (c) DHPLC chromatograms of PCR products of in vivo grown fg oocytes. n=9 chromatograms are superposed for H19, n= 8 for Mest/Peg1 and n=7 for Igf2R. (d) DHPLC chromatograms of PCR products of in vitro grown fg oocytes n=7 chromatograms are superposed: (→), unmethylated alleles; (↓), methylated alleles; (*), heteroduplexes.
Methylation analysis of imprinted Mest/Peg1 gene. (a) Genomic structure of the DMR studied. We amplified 22 CpGs of the Mest/Peg1 promoter region (accession no. AF017994; nt −201, +116 relative to transcriptional start). See Figure 1(a) for details. (b) DHPLC chromotograms of PCR products of control samples. See Figure 1(b) for details. (c) DHPLC chromotograms of PCR products of in vivo grown fg oocytes n=8 chromotograms are superposed. (d) DHPLC chromotograms of PLR products of in vitro grown fg oocytes. n=7 chromotograms are superposed. See Figure 1(d) for details.
Methylation analysis of imprinted Igf2R gene. (a) Genomic structure of the DMR studied. We amplified 13CpGs of the Igf2R within intronic DMR2 (accession no. L06446; nt 1857–2101). See Figure 1(a) for details. (b) DHPLC chromotograms of PCR products of control samples. See Figure 1(b) for details. (c) DHPLC chromotograms of PCR products of in vivo grown fg oocytes. n=7 chromotograms are superposed. (d) DHPLC chromotograms of PCR products of in vitro grown fg oocytes. n=8 chromotograms are superposed. For details see Figure 1(d).
After in vitro folliculogenesis, DHPLC profiles of PCR products from in vitro fg oocytes showed almost unmethylated alleles in six out of seven samples at the H19 locus (Figure 1d) and methylated alleles in six out of seven samples at the Mest/Peg1 locus (Figure 2d). In one (one out of seven) PCR product, a gain of allelic methylation and a loss or absence of allelic methylation were unexpectedly observed at the H19 and Mest/Peg1 loci, respectively. Such abnormal monoallelic profiles for H19 and Mest/Peg1 were different from multiallelic patterns that have been observed in granulosa cells (Figure 1, 2 and 3b). In contrast, at the Igf2R-R2 locus, DHPLC chromatograms showed profiles corresponding to almost methylated alleles in only one out of eight PCR products, whereas multiple peaks were found in seven out of eight samples (Figure 3d). These multiple peaks, consisting of homoduplex and heteroduplex formations were characteristics of heterogeneous samples with methylated alleles and unmethylated alleles, as confirmed by genomic bisulphite sequencing (not shown). However, since PCR amplifications were performed on pools of five to eight in vitro fg oocytes, we could not establish whether this heterogeneity affected individual fg oocytes (methylated and unmethylated alleles in the same fg oocyte) or the entire population of fg oocytes (fg oocytes with methylated alleles, fg oocytes with unmethylated alleles). Nevertheless, these results indicated, for the first time, the disruption (loss, absence or delay) of maternal methylation imprint establishment during in vitro folliculogenesis. The mechanisms that could affect maternal primary imprinting establishment as a consequence of in vitro culture remain elusive. We hypothesize that these dysregulations could be stochastic events at some imprinted loci as illustrated by improper de novo methylation at the H19 locus and improper demethylation at the Mest/Peg1 locus. Alternatively, these imprinting errors could be enhanced by in vitro cultivation at other specific imprinted loci, as exemplified by results at the Igf2R-R2 locus. The consequence of imprinting methylation errors for incomplete oocyte competence requires further investigation. However, it is tempting to speculate that such epigenetic abnormalities could be involved in subsequent abnormal development.12,13,14 Recently, complete oogenesis supporting development to full term after nuclear transfer and in vitro fertilization was successfully and efficiently accomplished by in vitro folliculogenesis using a 28-day in vitro culture system.15 This complete nuclear reprogramming was consistent with establishment of normal methylation imprinting at the Igf2R locus and illustrates the critical role of culture procedures on subsequent epigenotype and phenotype. Although, in vitro folliculogenesis will enable important progress in different fields of biology and medicine, these results highlight the need for improving in vitro culture conditions and developing biomarkers to identify complete nuclear competence.
References
Smitz JE, Cortvrindt RG : The earliest stages of folliculogenesis in vitro. Reproduction 2002; 123: 185–202.
Obata Y, Kono T : Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem 2002; 277: 5285–5289.
Reik W, Dean W, Walter J : Epigenetic reprogramming in mammalian development. Science 2001; 293: 1089–1092.
Surani MA : Imprinting and the initiation of gene silencing in the germ line. Cell 1998; 93: 309–312.
Kono T, Obata Y, Yoshimzu T, Nakahara T, Carroll J : Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat Genet 1996; 13: 91–94.
Dean W, Bowden L, Aitchison A et al: Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 1998; 125: 2273–2282.
Khosla S, Dean W, Brown D, Reik W, Feil R : Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod 2001; 64: 918–926.
Cortvrindt R, Smitz J, Van Steirteghem AC : In-vitro maturation, fertilization and embryo development of immature oocytes from early preantral follicles from prepuberal mice in a simplified culture system. Hum Reprod 1996; 11: 2656–2666.
Kerjean A, Vieillefond A, Thiounn N et al: Bisulfite genomic sequencing of microdissected cells. Nucleic Acids Res 2001; 29: e106.
Couvert P, Poirier K, Carrié A et al: A DHPLC-based method for DNA methylation analysis of differential methylated regions from imprinted genes. Bio Techniques, 2003; 34: 356–362.
Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM : Methylation dynamics of imprinted genes in mouse germ cells. Genomics 2002; 79: 530–538.
DeBaun MR, Niemitz EL, McNeil DE et al: Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith–Wiedemann syndrome with cancer and birth defects. Am J Hum Genet 2002; 70: 604–611.
Young LE, Sinclair KD, Wilmut I : Large offspring syndrome in cattle and sheep. Rev Reprod 1998; 3: 155–163.
Young LE, Fernandes K, McEvoy TG et al: Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet 2001; 27: 153–154.
Obata Y, Kono T, Hatada I : Maturation of mouse fetal germ cells in vitro. Nature 2002; 418: 497.
Acknowledgements
This work was supported by a grant from the ‘la Société Générale’. We thank Susan Saint-Just for reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kerjean, A., Couvert, P., Heams, T. et al. In vitro follicular growth affects oocyte imprinting establishment in mice. Eur J Hum Genet 11, 493–496 (2003). https://doi.org/10.1038/sj.ejhg.5200990
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200990
Keywords
This article is cited by
-
Intragenic sequences in the trophectoderm harbour the greatest proportion of methylation errors in day 17 bovine conceptuses generated using assisted reproductive technologies
BMC Genomics (2018)
-
Live birth rate after human chorionic gonadotropin priming in vitro maturation in women with polycystic ovary syndrome
Journal of Ovarian Research (2018)
-
In vitro fertilization (IVF) in mammals: epigenetic and developmental alterations. Scientific and bioethical implications for IVF in humans
Biological Research (2015)
-
Variable imprinting of the MEST gene in human preimplantation embryos
European Journal of Human Genetics (2013)
-
Health of IVM children
Journal of Assisted Reproduction and Genetics (2011)