Abstract
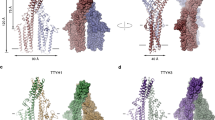
Ion-coupled membrane-transport proteins, or secondary transporters, comprise a diverse and abundant group of membrane proteins that are found in all organisms. These proteins facilitate solute accumulation and toxin removal against concentration gradients using energy supplied by ion gradients across membranes. NhaA is a Na+/H+ antiporter of relative molecular mass 42,000, which is found in the inner membrane of Escherichia coli, and which has been cloned and characterized1,2. NhaA uses the H+ electrochemical gradient to expel Na+ from the cytoplasm, and functions primarily in the adaptation to high salinity at alkaline pH1,2. Most secondary transporters, including NhaA3, are predicted to have 12 transmembrane helices. Here we report the structure of NhaA, at 7?Å resolution in the membrane plane and at 14?Å vertical resolution, determined from two-dimensional crystals4 using electron cryo-microscopy. The three-dimensional map of NhaA reveals 12 tilted, bilayer-spanning helices. A roughly linear arrangement of six helices is adjacent to a compact bundle of six helices, with the density for one helix in the bundle not continuous through the membrane. The molecular organization of NhaA represents a new membrane-protein structural motif and offers the first insights into the architecture of an ion-coupled transport protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Padan,E. et al. The molecular mechanism of regulation of the NhaA Na+/H+ antiporter of Escherichia coli, a key transporter in the adaptation to Na+ and H+. Novartis Found. Symp. 221, 183–196 (1999).
Padan,E. & Schuldiner,S. in Handbook of Biological Physics (eds Konings, W. N., Kaback, H. R. & Lolkema, J. S.) 501–531 (Elsevier, Oxford, 1996).
Rothman,A., Padan,E. & Schuldiner,S. Topological analysis of NhaA, a Na+/H+ antiporter from Escherichia coli. J. Biol. Chem. 271, 32288–32292 (1996).
Williams,K. A. et al. Projection structure of NhaA, a secondary transporter from Escherichia coli, at 4.0?Å resolution. EMBO J. 18, 3558–3563 (1999).
Blakely,R. D., De Felice,L. J. & Hartzell,H. C. Molecular physiology of norepinephrine and serotonin transporters. J. Exp. Biol. 196, 263–281 (1994).
Schloss,P. & Williams,D. C. The serotonin transporter: a primary target for antidepressant drugs. J. Psychopharmacol. 12, 115–121 (1998).
Wright,E. M. Glucose galactose malabsorption. Am. J. Physiol. 275, 879–882 (1998).
Saier,M. H. Jr et al. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12, 265–274 (1998).
Kaback,H. R. & Wu,J. From membrane to molecule to the third amino acid from the left with a membrane transport protein. Q. Rev. Biophys. 30, 333–364 (1997).
Iwata,S., Ostermeier,C., Ludwig,B. & Michel,H. Structure at 2.8?Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376, 660–669 (1995).
Auer,M., Scarborough,G. A. & Kühlbrandt,W. Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature 392, 840–843 (1998).
Zhang,P. et al. Structure of the calcium pump from sarcoplasmic reticulum at 8?Å resolution. Nature 392, 835–839 (1998).
Henderson,R. et al. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 213, 899–929 (1990).
Grigorieff,N., Ceska,T. A., Downing,K. H., Baldwin,J. M. & Henderson,R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259, 393–421 (1996).
Havelka,W. A., Henderson,R. & Oesterhelt,D. Three-dimensional structure of halorhodopsin at 7?Å resolution. J. Mol. Biol. 247, 726–738 (1995).
Doyle,D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998).
Chang,G., Spencer,R. H., Lee,A. T., Barclay,M. T. & Rees,D. C. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science 282, 2220–2226 (1998).
Walz,T. et al. The three-dimensional structure of aquaporin-1. Nature 387, 624–627 (1997).
Cheng,A. et al. Three-dimensional organization of a human water channel. Nature 387, 627–630 (1997).
Li,H., Lee,S. & Jap,B. K. Molecular design of aquaporin-1 water channel as revealed by electron crystallography. Nature Struct. Biol. 4, 263–265 (1997).
Rothman,A., Gerchman,Y., Padan,E. & Schuldiner,S. Probing the conformation of NhaA, a Na+/H+ antiporter from Escherichia coli, with trypsin. Biochemistry 36, 14572–14576 (1997).
Unwin,N. Acetylcholine receptor in the open state. Nature 373, 37–43 (1995).
Subramaniam,S. et al. Protein conformational changes in the bacteriorhodopsin photocycle. J. Mol. Biol. 287, 145–161 (1999).
Peruse,E., Courts,D. M. & Cull,L. G. Structural rearrangements underlying K+ channel activation gating. Science 285, 73–78 (1999).
Wang,D. N. & Kühlbrandt,W. High-resolution electron crystallography of light-harvesting chlorophyll a/b-protein complex in three different media. J. Mol. Biol. 217, 691–699 (1991).
Henderson,R. et al. Structure of purple membrane from Halobacterium halobium: recording, measurement and evaluation of electron micrographs at 3.5?Å resolution. Ultramicroscopy 19, 899–929 (1986).
Crowther,R. A., Henderson,R. & Smith,J. M. MRC image processing programs. J. Struct. Biol. 116, 9–16 (1996).
Shaw,P. J. & Hills,G. J. Tilted specimen in the electron microscope: a simple specimen holder and the calculation of tilt angles from crystalline specimens. Micron 12, 279–282 (1981).
Unger,V. M. & Schertler,G. F. X. Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys. J. 68, 1776–1786 (1995).
Jones,T. A., Zou,J. Y., Cowan,S. W. & Kjeldgaard,M. Improved methods for building protein models in electron density maps. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
I thank W. Kühlbrandt for his support and encouragement throughout this project, D. Mills for assistance with the JEOL 3000 SFF, V. Unger for advice on image processing, U. Geldmacher-Kaufer for technical assistance, and S. Schuldiner, E. Padan and A. Rothman for their assistance with NhaA biochemistry during the early stages of this work. This work was supported by the Deutsche Forschungsgesellshaft.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
Supplementary Information (PDF 59 kb)
Rights and permissions
About this article
Cite this article
Williams, K. Three-dimensional structure of the ion-coupled transport protein NhaA. Nature 403, 112–115 (2000). https://doi.org/10.1038/47534
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/47534
This article is cited by
-
Crystal structure of the Na+/H+ antiporter NhaA at active pH reveals the mechanistic basis for pH sensing
Nature Communications (2022)
-
Role of the Group 2 Mrp sodium/proton antiporter in rapid response to high alkaline shock in the alkaline- and salt-tolerant Dietzia sp. DQ12-45-1b
Applied Microbiology and Biotechnology (2018)
-
Integrating mass spectrometry with MD simulations reveals the role of lipids in Na+/H+ antiporters
Nature Communications (2017)
-
Living long and ageing well: is epigenomics the missing link between nature and nurture?
Biogerontology (2016)
-
A two-domain elevator mechanism for sodium/proton antiport
Nature (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.