Abstract
In neurons, DNA is prone to free radical damage, although repair mechanisms preserve the genomic integrity. However, activation of the DNA repair system, poly(ADP-ribose) polymerase (PARP-1), is thought to cause neuronal death through NAD+ depletion and mitochondrial membrane potential (Δψm) depolarization. Here, we show that abolishing PARP-1 activity in primary cortical neurons can either enhance or prevent apoptotic death, depending on the intensity of an oxidative stress. Only in severe oxidative stress does PARP-1 activation result in NAD+ and ATP depletion and neuronal death. To investigate the role of PARP-1 in an endogenous model of oxidative stress, we used an RNA interference (RNAi) strategy to specifically knock down glutamate-cysteine ligase (GCL), the rate-limiting enzyme of glutathione biosynthesis. GCL RNAi spontaneously elicited a mild type of oxidative stress that was enough to stimulate PARP-1 in a Ca2+-calmodulin kinase II-dependent manner. GCL RNAi-mediated PARP-1 activation facilitated DNA repair, although neurons underwent Δψm loss followed by some apoptotic death. PARP-1 inhibition did not prevent Δψm loss, but enhanced the vulnerability of neurons to apoptosis upon GCL silencing. Conversely, mild expression of PARP-1 partially prevented to GCL RNAi-dependent apoptosis. Thus, in the mild progressive damage likely occur in neurodegenerative diseases, PARP-1 activation plays a neuroprotective role that should be taken into account when considering therapeutic strategies.
Similar content being viewed by others
Main
Neurons are prone to acute oxidative stress.1, 2, 3, 4 Furthermore, accumulation of DNA strand breaks is a contributing factor to neurodegeneration during oxidative stress and other conditions such as exposure to alkylating agents5, 6 and aging.7 To repair damaged DNA, most eukaryotic cells – except yeast – express poly(ADP-ribose) polymerase-1 ((PARP-1); EC 2.4.2.30),8 a nuclear enzyme that rapidly binds to DNA strand breaks leading to the formation of long, branched poly(ADP-ribose) polymers using NAD+ as substrate. The resulting negatively charged PARP-1 is subsequently dissociated from DNA ends, facilitating the DNA repair process.8, 9, 10
Activation of PARP-1 secondary to the acute exposure of neurons and other cell types to pro-oxidant species, such as nitric oxide, peroxynitrite or hydrogen peroxide (H2O2)11, 12, 13 has been shown to lead to NAD+ depletion and energetic failure, ultimately resulting in cellular death. Moreover, such activation of PARP-1 has been shown to be associated with mitochondrial impairment and apoptotic death, at least in models of oxygen and glucose deprivation in neurons.14, 15 This has led to the suggestion that PARP-1 inhibitors might be neuroprotective.15, 16, 17 However, in a model of apoptosis dependent on the use of an essential medium lacking in insulin, it has recently been demonstrated that in neurons18 DNA repair mechanisms contribute to essential housekeeping functions, maintaining the neuronal genome in a healthy status. Furthermore, these authors showed that increasing the intensity of the apoptotic stimulus overrides the DNA repair process.18 We surmised that the discrepancies in the above studies might be owing to the degree and nature of the insult utilized and that the response to drastic exogenous stimuli might be qualitatively different from an endogenous progressive stress that resembles more closely the process of chronic neurodegeneration.
To clarify this, we investigated the role played by PARP-1 activity in neuronal death/survival in response to oxidative stress of different intensities. We found that only in severe oxidative stress, induced by a high concentration of H2O2, did activation of PARP-1 result in NAD+ and ATP depletion and cell death. In contrast, mild oxidative stress, induced either by exogenous modulation of glutathione redox status or by endogenously interfering with glutathione biosynthesis, led to neuroprotection by PARP-1.
Results
PARP-1 mediates death or survival in primary cortical neurons depending on the intensity of the oxidative insult
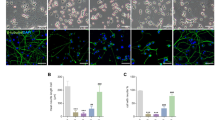
We first induced oxidative stress by inhibiting glutathione biosynthesis in primary cortical neurons with L-buthionine sulfoximine (L-BSO), an inhibitor of glutamate-cysteine ligase (GCL),19 the rate-limiting enzyme of glutathione biosynthesis.20 Glutathione concentrations decreased in a time-dependent manner (Figure 1a), leading to accumulation of poly (ADP-ribose) polymers (PAR) (Figure 1b) and moderate apoptotic death (annexin V+/7-amino-actinomycin D (7-AAD−), as assessed by flow cytometry; Figure 1c) 24 h after L-BSO treatment. Inhibition of PARP-1 activity with 3-aminobenzamide (ABA) or 5-aminoisoquinolinone (AIQ) prevented PAR accumulation (Figure 1b) and enhanced neuronal apoptotic death (Figure 1c) caused by L-BSO. As our data appear to conflict with other studies,15, 16, 21 we wondered whether the intensity of oxidative insult might explain our findings. Neurons were incubated with 50 or 500 μM H2O2 to produce mild or severe oxidative stress, respectively. The intensity of oxidative stress was confirmed by determining the degree of glutathione oxidation that, although transient, was severe at 500 μM and mild at 50 μM H2O2 (Figure 1d). Likewise, both stimuli decreased NAD+ concentrations (Figure 1e), consistent with an activation of PARP-1;8 however, only the severe oxidative insult decreased ATP concentrations (Figure 1f). Inhibition of PARP-1 with ABA or AIQ fully restored NAD+ concentrations (Figure 1e) and significantly protected against the decrease in ATP (Figure 1f). We next investigated the function of PARP-1 in both severe (500 μM H2O2) and mild (50 μM H2O2) oxidative stress. We found that treatment of primary neurons with the severe insult (500 μM H2O2) triggered apoptotic death at 1 h, whereas it was necessary to incubate for 18 h in order to observe cell death after the mild insult (50 μM H2O2) (Figure 1g). Inhibition of PARP-1 activity with ABA or AIQ either prevented (Figure 1g, left) or enhanced (Figure 1g, right) apoptotic death caused by the severe or mild oxidative insults, respectively. To confirm this, we inhibited PARP-1 by RNA interference (RNAi) using a pSuper-neo/gfp-based22 small hairpin RNA (shRNA) approach.14 Non-transfected neurons profusely expressed nuclear PARP-1 (Figure 1h, middle and right); expression of PARP-1 shRNA decreased PARP-1 protein without altering neuronal survival, as reflected by the intact nuclei in the green fluorescent protein (GFP+) neurons (Figure 1h, left and middle). Assessments of GFP+ apoptotic (annexin V+/7-AAD−) neurons by flow cytometry confirmed that inhibition of PARP-1, while not affecting neuronal survival, either prevented or enhanced neuronal death caused by severe or mild oxidative insults, respectively (Figure 1i).
PARP-1 mediates death or survival in primary cortical neurons depending on the intensity of the oxidative insult. (a) L-BSO, 0.5 mM time-dependently decreased total glutathione concentrations in neurons. (b) Incubation of neurons with L-BSO for 24 h triggered poly(ADP-ribosyl)ation mediated by PARP-1, as judged by Western blotting against poly(ADP-ribose) (PAR) in the absence (control) or presence of PARP-1 inhibitors ABA, 0.5 mM and AIQ, 50 μM. (c) Incubation of neurons with L-BSO caused, after 24 h, an increase in apoptotic neuronal death (annexin V+/7-AAD−) that was enhanced by PARP-1 inhibition. (d) Hydrogen peroxide (H2O2) triggered mild (50 μM) or severe (500 μM) glutathione oxidation in neurons. (e) The mild oxidative insult decreased NAD+ concentrations by approximately 50%, whereas the severe insult caused NAD+ depletion after 1 h of incubation; however, PARP-1 inhibition with ABA or AIQ fully prevented the decrease in NAD+ concentrations. (f) Severe, but not mild oxidative insult promoted ATP depletion, which was largely prevented by PARP-1 inhibition with ABA or AIQ. (g) Inhibition of PARP-1 with ABA or AIQ prevented (left) or increased (right) apoptotic death of neurons treated with severe or mild oxidative insults, respectively. (h) Transfection of primary neurons with a vector expressing PARP-1 shRNA efficiently depleted PARP-1 protein, as revealed by the lack of PARP-1 staining in GFP+ cells (left and middle, cells indicated by arrows); the viability of these neurons was not altered, as the integrity of their nuclei stained with DAPI was intact (right). (i) PARP-1 shRNA alone did not confer toxicity but either prevented (left) or enhanced (right) apoptosis in GFP+ neurons subjected to severe or mild oxidative insults, respectively. Tub, tubulin (used as loading control). (Bar=60 μm). *P<0.05 as compared with the corresponding – L-BSO in (c), 0 min in (d), 0 μM H2O2 in (e, f, g and i). #P<0.05 as compared with the corresponding. None in C, 50 μM H2O2 in (d), None in (f) and (g) control (luciferase) shRNA in (i)
Interference with glutathione biosynthesis in primary neurons by RNAi spontaneously induces free radical production and PARP-1-mediated DNA repair
We next sought to confirm our results using a further model of mild and endogenous neuronal oxidative stress. We designed a pSuper-neo/gfp22 vector-derived shRNA strategy to target the catalytic subunit of GCL. The effectiveness of this GCL shRNA was first confirmed in the cell line human embryonic kidney 293T (HEK293T) –which shows high transfection efficiencies–, where glutathione concentrations were reduced (Figure 2a, left). Furthermore, this effect was specific, as it prevented the protein expression of wild-type GCL full-length cDNA, but not that of site-directed mutated GCL cDNA refractory to the shRNA sequence (Figure 2a, right) (see, also23). Transfection of primary cortical neurons with this GCL shRNA (displaying GFP fluorescence) efficiently decreased endogenous GCL (Figure 2b) and spontaneously elicited oxidative stress, as judged by rhodamine-123 (Rh-123) fluorescence (Figure 2c). GCL RNAi-induced oxidative stress was confirmed with hydroethidine (HE), which displays red fluorescence when oxidized (Figure 2d, right panels). The free radical scavenger tempol (100 μM) prevented GCL RNAi-mediated HE oxidation, whereas H2O2 treatment (500 μM) strongly oxidized HE (Figure 2d). Furthermore, we found PARP-1 to be activated in this model, as GCL shRNA (Figure 2e, lower panels), but not control shRNA (Figure 2e, upper panels), promoted poly(ADP-ribosyl)ation, as judged by PAR immunostaining in GFP+ neurons (control shRNA, 18.7±2.1% GFP+ neurons; GCL shRNA, 75.8±8.3% GFP+ neurons; ∼500 cells counted for each condition). In agreement with this, GCL silencing increased bromodeoxyuridine (BrdU) incorporation into the nucleus of GFP+ cells, as assessed by flow cytometry (Figure 2f); this effect was fully prevented by scavenging free radicals with tempol and by inhibiting PARP-1 activity with ABA and AIQ (Figure 2f).
Interference with glutathione biosynthesis in primary neurons by RNAi spontaneously induces free radical production and PARP-1-mediated DNA repair. (a) GCL shRNA decreases glutathione concentrations in HEK293T cells 24 h after transfection (left); site-directed mutagenesis of GCL with silent point mutations at the third codon-base within the shRNA-target sequence (GCLmut) resulted in the expression of proteins that were refractory to the shRNA-mediated protein degradation, as assessed in HEK293T cells (right). (b) Transfection of primary neurons with GCL shRNA (displaying GFP+ fluorescence) efficiently depleted GCL (top) after 24 h. (c) Transfection of primary neurons (identified as Map-2-immunopositive cells; middle) with GCL shRNA (displaying GFP+ fluorescence) generated, after 24 h, oxygen free radicals, as judged by the ability of GFP+ neurons to oxidize dihydrorhodamine-123 to Rh-123 (bottom); however, GFP−/Map-2+ neurons failed to oxidize dihydrorhodamine-123. (d) GCL silencing promoted HE fluorescence; the specificity of the probe was tested by tempol (100 μM), which prevented, and H2O2 (500 μM), which enhanced HE fluorescence. (e) GCL silencing triggered an increase in cellular poly(ADP-ribosyl)ation, as revealed by anti-PAR immunocytochemistry of GFP+ neurons when comparing control shRNA (upper panels) with GCL shRNA (lower panels). (f) GCL silencing increased BrdU incorporation into the DNA of GFP+ cells, which was prevented by tempol (100 μM) and by PARP-1 inhibitors ABA (0.5 mM) and AIQ (50 μM). Tub, tubulin (used as loading control). (Bars=60 μm). *P<0.05 as compared with the corresponding Control (luciferase) shRNA in (a and f). #P<0.05 as compared with the corresponding None in (f)
Knock down of GCL by RNAi triggers neuronal apoptotic death through the intrinsic (mitochondrial) pathway
Silencing of neuronal GCL spontaneously induced apoptotic death, as revealed by immunocytochemistry (TUNEL; Figure 3a,b) or by flow cytometry (annexin V+/7AAD−) (Figures 3b,c) in GFP+ cells. We then assessed the proportion of GFP+ annexin V+/7AAD− apoptotic neurons after transfection with the GCL-targeted shRNA in the presence of increasing concentrations of a battery of caspase activity inhibitors (Calbiochem). As shown in Figure 4a (left and middle upper panels), ZVAD-fmk and ZDEVD-fmk, which are non-specific caspase inhibitors that do not distinguish among its subtypes, prevented the death of GFP+ neurons. Incubation of neurons with ZDQMD-fmk, a compound reported to be a highly specific inhibitor of caspase-3 activity, also prevented apoptotic death of GFP+ neurons (Figure 4a, right upper panel). As caspase-3 can be activated by the mitochondrial-independent caspase-2, -8 or the mitochondrial-dependent caspase-9, we next used inhibitors reported to inhibit those caspase subtypes specifically. As shown in Figure 4a (left and middle lower panels), ZIETD-fmk and ZVDVAD-fmk, inhibitors of caspases-8 and -2, respectively, did not prevent neuronal death even at the highest concentrations used (100 μM). In contrast, ZLEHD-fmk, a potent and specific inhibitor of caspase-9, prevented apoptotic death of GFP+ neurons in a concentration-dependent manner (Figure 4a, right lower panel). These results suggest that apoptotic neuronal death upon GCL silencing took place via the intrinsic pathway, that is, involving the activity of caspase-9 and -3. This was further confirmed by establishing, using flow cytometry, that the proportion of active caspase-3 was significantly increased in GFP+ neurons (Figure 4b, c), reinforcing the notion of mitochondrial involvement in the cell death pathway after GCL silencing. Since, under certain circumstances, caspase-3 has been reported to cleave PARP-1,24 we next investigated whether the activation of caspase-3 observed in our experiments was associated with PARP-1 cleavage. We found that GCL shRNA did not produce PARP-1 cleavage, as judged by flow cytometry (Figure 4d). Positive controls using etoposide (1 μM) validated this experimental approach, as neurons incubated with this compound showed sharp increases in both caspase-3 activity (Figure 4b, right panel) and PARP-1 cleavage (Figure 4d, right panel).
Knockdown of glutathione biosynthesis by RNAi triggers neuronal apoptotic death. (a) Primary neurons expressing the shRNA against GCL, but not those expressing the control (luciferase) shRNA, were TUNEL-positive 48 h after transfection. (b) Cell death was confirmed by the analysis of annexin V+/7AAD− neurons by flow cytometry within the GFP+ cells (left), giving rise to values that were similar to those obtained by counting the proportion of TUNEL+/GFP+ neurons in the microphotographs (right). (c) Flow cytometric results of a typical experiment in primary cortical neurons showing increased annexin V+/7-AAD− neurons by GCL shRNA. (Bar=60 μm). *P<0.05 as compared with corresponding control (luciferase) shRNA
Knockdown of glutathione biosynthesis by RNAi triggers neuronal apoptotic death through the intrinsic (mitochondrial) pathway. (a) GFP+ annexin V+/7AAD− apoptotic neurons after transfection with GCL shRNA were assessed after 24 h in the presence of increasing concentrations of inhibitors of caspase activity. ZVAD-fmk and ZDEVD-fmk (nonspecific caspase inhibitors that do not distinguish among its subtypes) prevented the death of GFP+ neurons; ZDQMD-fmk, a highly specific inhibitor of caspase-3 activity, prevented apoptotic death of GFP+ neurons; ZIETD-fmk and ZVDVAD-fmk (inhibitors of caspases-8 and -2, respectively) did not prevent neuronal death even at the highest concentrations used (100 μM); ZLEHD (a potent and specific inhibitor of caspase-9) prevented apoptotic death of GFP+ neurons in a concentration-dependent manner. (b) Flow cytometric diagram of a typical experiment of active caspase-3-phycoerythrine conjugate (PE) staining in control and GCL shRNA-treated primary cortical neurons, either in the absence or presence of 1 μM etoposide. (c) Quantitative analysis showing that GCL shRNA increased the proportion of active caspase-3-PE-stained GFP+, but not GFP−, neurons. (d) Flow cytometric diagram of a typical experiment of cleaved PARP-1-PE staining in control and GCL shRNA-treated primary cortical neurons, either in the absence or presence (positive control) of 1 μM etoposide. *P<0.05 as compared with corresponding Control (luciferase) shRNA
PARP-1 inhibition enhances apoptotic death without altering the early Δψm loss triggered by GCL knock-down
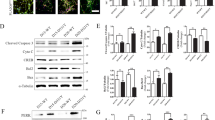
In order to assess directly the effect of GCL knock-down on mitochondria, we determined Δψm and apoptotic death in primary neurons. We found that GCL silencing triggered mitochondrial membrane potential (Δψm) loss (Figure 5a) and neuronal apoptotic death (Figure 5b) that were due to the action of free radicals, as tempol fully prevented both parameters (Figure 5a, b, c). Furthermore, neuronal death was preceded by Δψm loss after GCL knockdown (compare data at 16 h; Figure 5a, b), suggesting that free radical-mediated mitochondrial damage would be an early feature following GCL knockdown. Pharmacological inhibition of PARP-1 activity with ABA or AIQ did not alter the Δψm loss (Figure 5d), but it significantly enhanced apoptotic neuronal death by GCL knockdown (Figure 5e). To investigate this further, we decided to silence PARP-1 by RNAi in GCL shRNA neurons. As shown in Figure 5f, PARP-1 disruption in GCL-deficient neurons (revealed as GFP+/PARP-1− cells) showed nuclei condensation by DAPI staining, suggesting apoptotic death; however, PARP-1-expressing neurons (GFP−/PARP-1+) displayed intact nuclei. To confirm this effect, we quantified the proportion of GFP+ apoptotic (annexin V+/7AAD−) neurons by flow cytometry. As shown in Figure 5g, PARP-1 silencing did not affect, per se, neuronal survival, but synergistically enhanced apoptotic death by GCL deficiency. These data were confirmed using L-BSO in PARP-1 shRNA cells (Figure 5h). To directly test neuroprotection by PARP-1, GCL shRNA-expressing neurons were transfected with increasing amounts of a pEXPRESS vector encoding the full-length PARP-1 cDNA. PARP-1 overexpression triggered apoptotic death only at concentrations higher than 0.3 μg of DNA/103 cells (Figure 5i). This is consistent with the results of other authors showing neuronal death upon excessive activation of PARP-1.25, 26 Furthermore, the expression of PARP-1 at non-toxic concentrations (0.3 μg/103 cells) partially prevented GCL RNAi-mediated apoptotic death (Figure 5j). Finally, we investigated whether translocation of apoptosis-inducing factor (AIF) from mitochondria to the nucleus – a typical marker of PARP-1-mediated apoptosis under certain circumstances25, 27, 28 – would occur in the GCL RNAi mild oxidative stress model. Immunofluorescence experiments (Figure 6a) revealed that AIF remained in the cytoplasm, preferentially within the perinuclear area, during GCL knockdown (see GFP+ neurons in Figure 6a).
PARP-1 inhibition enhances apoptotic death but does not alter the early Δψm loss triggered by GCL knockdown. (a) Δψm was significantly decreased 16 h after transfection of primary neurons with GCL shRNA; tempol (100 μM) fully prevented the effects caused by GCL silencing. (b) At 16 h after GCL shRNA treatment, no apoptotic cell death was observed within the GFP+ neurons. (c) Tempol concentration-dependently prevented apoptotic death of GFP+ neurons 24 h after GCL shRNA treatment, revealing that 100 μM of the scavenger was fully effective. (d) PARP-1 inhibition with ABA (0.5 mM) or AIQ (50 μM) did not affect the Δψm loss caused by GCL silencing. (e) PARP-1 inhibition with ABA (0.5 mM) or AIQ (50 μM) enhanced apoptotic death caused by GCL silencing. (f) Co-transfection of primary neurons with PARP-1 and GCL shRNAs (GFP+/PARP-1− cells) resulted in nuclei condensation, as revealed by DAPI immunostaining. (g) Flow cytometric analyses within the GFP+ apoptotic (annexin V+/7AAD−) neurons confirm that GCL silencing alone promoted apoptotic death, an effect that was synergistically enhanced by PARP-1 co-depletion. (h) Flow cytometric analyses within the GFP+ apoptotic (annexin V+/7AAD−) neurons confirm that, 24 h after L-BSO (0.5 mM) treatment, apoptotic death occurred, an effect that was synergistically enhanced by PARP-1 shRNA. (i) Transfection of primary neurons with pEXPRESS-PARP-1 revealed, after 24 h, a concentration-dependent increase in apoptotic death as evidenced by flow cytometry (annexin V+/7AAD− within GFP+ neurons). pEXPRESS-PARP-1 was coexpressed with GFP-encoding pd2EGFP-N1 plasmid vector at a 3 : 1 ratio. (j) Expression of PARP-1 in neurons using non-toxic concentrations of p-EXPRESS-PARP-1 (0.3 μg/106 cells) partially prevented GCL shRNA-dependent apoptotic death, as assessed by flow cytometry. (Bar=30 μm). *P<0.05 as compared with corresponding control (luciferase) shRNA in (a, b, d and e); with – GCL shRNA in (g) and (j); or with – L-BSO in (h). #P<0.05 as compared with corresponding none in (e); or with control (luciferase) shRNA in (g, h and j)
PARP-1 activation and neuroprotection after GCL knockdown is mediated by CaMK-II. (a) Immunofluorescence detection of AIF (using a PE conjugate; red fluorescence) confirms the preferential perinuclear localization of AIF both in GCL shRNA-expressing (GFP+) and in GCL shRNA-not expressing (GFP−) cortical neurons (Bar=15 μm). (b and c), Incubation of neurons in a Ca2+-chelating medium (Ca2+-free medium containing 100 μM EDTA) or in the presence of CaMK-II inhibitor, KN-62 (5 μM), prevented both GCL shRNA- (b) or L-BSO-induced (c) poly(ADP-ribosyl)ation, as revealed by anti-PAR immunocytochemistry of GFP+ neurons (b) or Western blotting (c). (d and e), Ca2+ chelation (Ca2+-free media containing 100 μM EDTA or 10 μM BAPTA), or inhibition of CaMK-II (5 μM KN-62) enhanced the apoptotic death induced by GCL shRNA (d) or L-BSO (e)
GCL knockdown activates PARP-1 through Ca2+-calmodulin-dependent protein kinase II (CaMK-II) in cortical neurons
To ascertain the mechanism responsible for RNAi-mediated PARP-1 activation, we focused on CaMK-II, as it has recently been reported that CaMK-II controls PARP-1 activation by promoting the dissociation of PARP-1 from kinesin superfamily protein 4 (KIF4).29 We first investigated poly-ADP ribosylation using an antibody against poly(ADP) ribosyl (PAR) polymers (Figure 6b, c). Inhibition of glutathione biosynthesis with either GCL shRNA or L-BSO increased PAR accumulation, as judged by immunofluorescence (Figure 6b, left panels) or Western blotting (Figure 6c), respectively. Both chelation of Ca2+ with EDTA or inhibition of CaMK-II activity with KN-6229 prevented PAR accumulation (Figure 6b, middle panels; Figure 6c). Finally, we assessed whether the CaMK-II pathway mediated the neuroprotective effect of PARP-1 against mild oxidative insults. Chelation of Ca2+ with EDTA or BAPTA (1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetra(acetoxymethyl) ester, membrane-permeable form; Calbiochem), or inhibition of CaMK-II with KN-62 enhanced the apoptotic death induced by treatment of primary neurons with either GCL shRNA or L-BSO (Figures 6d and e, respectively).
Discussion
Here we show that decreasing glutathione concentrations with L-BSO, an inhibitor of GCL, triggered a moderate degree of apoptotic death and showed evidence of PARP-1 activation. Inhibition of PARP-1 in L-BSO-treated neurons enhanced, rather than decreased, apoptotic cell death. This result is unexpected, as there is a widely held belief that stimulation of PARP-1 activity is associated with neuronal death.15, 17, 30 This may occur as a consequence of NAD+ depletion, energetic failure and Δψm loss, and has been reported to occur following pro-oxidant insult11, 12, 13, 31 Because of this it has been suggested that PARP-1 inhibitors might be protective against neuronal damage15, 16 and this has recently been demonstrated in the rat in a model of cerebral infarction induced by middle cerebral artery occlusion.21 As our data appear to conflict with such studies, we wondered whether the intensity of oxidative insult might explain our findings.
Neurons were exposed to mild or severe oxidative stress, respectively, using different concentrations of H2O2. The severity of the oxidative stress was determined by assessing the glutathione redox status. Both treatments decreased NAD+ concentrations, confirming the activation of PARP-1.8 However, the concentration of NAD+ decreased only by 50% following mild oxidative insult, whereas there was almost complete depletion following severe oxidative stress; this was further corroborated by ATP values, which were decreased only by the severe oxidative insult. Our data showing prevention of ATP decrease by PARP-1 inhibition confirms the widely held notion that PARP-1 activation after severe oxidative insult causes energy failure.11, 12, 13, 31 Moreover, we found that both severe and mild oxidative stress triggered apoptotic neuronal death. Surprisingly, inhibition of PARP-1 (either by pharmacological inhibition or by RNAi strategy), either prevented or enhanced apoptotic death caused by the severe or mild oxidative insults, respectively. These results indicate that the effect of PARP-1 on neurodegeneration depends on the degree of oxidative damage.
We have recently developed a technique, based on the disruption of GCL activity using shRNA, which induces progressive neurodegeneration.23 This strategy is likely to mimic mild, endogenous oxidative stress, such as that which presumably occurs in chronic neurodegenerative diseases.5, 6 First, we confirmed that the expression of this GCL shRNA in primary neurons decreased endogenous GCL, spontaneously elicited oxidative stress and activated PARP-1. Silencing GCL increased BrdU incorporation into the nucleus, an effect that was fully prevented by scavenging free radicals with tempol or by inhibiting PARP-1 activity. These results suggest that the endogenous free radical generation in this neuronal model of oxidative stress induces DNA damage and repair by PARP-1 activity. Moreover, GCL silencing spontaneously induced apoptotic death via the mitochondrial – intrinsic – pathway, confirming the involvement of mitochondria in oxidative stress-mediated neurodegeneration.32
These mitochondrial changes are an early occurrence and probably lead to initiation of caspase-dependent cell damage, as has previously been suggested.33 The mitochondrial changes are independent of PARP-1 activation, which occurs in parallel, but their consequences can be modified by it, as inhibition of PARP-1 did not affect the loss of Δψm caused by GCL deficiency but still enhanced the neurotoxicity. Under more severe circumstances, caspase-3 has been shown to cleave PARP-1.24 However, GCL knocked-down neurons showed intact (not cleaved) PARP-1 despite having increased caspase-3 activity. Thus, we propose that in this kind of mild progressive neuronal damage there may be formation of free radicals leading to mitochondrial damage and, unrelated to it, PARP-1 activation acting as a defense mechanism; indeed our results show that moderate expression of PARP-1 partially protected GCL deficient neurons against apoptosis. This sequence of events clearly differs from the more dramatic situation in which excessive PARP-1 activation leads to energy depletion and cell death via a mechanism that is caspase-independent25 but may involve translocation from the mitochondria to the nucleus of the AIF.25, 27 In our experiments, GCL deficiency did not alter the subcellular localization of AIF, which remained within the perinuclear area, consistent with mitochondrial localization of this transcription factor.28 This situation may account for the results obtained with PARP-1 inhibitors in the severe model of cerebral infarction in the rat.21 Interestingly, it has recently been suggested that PARP-1 supports the survival of developing neurons through a mechanism involving CaMK-II-dependent dissociation from PARP-1 of the KIF4, whose C-terminal domain suppresses the activity of PARP-1.29 Here, we confirmed the involvement of CaMK-II in PARP-1-dependent neuroprotection during the low level stress induced by GCL knockdown. Our results indicate that caution should be exercised when considering the use of PARP-1 inhibitors in chronic neurodegenerative diseases.
Materials and Methods
Plasmid construction for transient expression of shRNAs
GCL and PARP-1 knockdown was achieved by RNAi using a vector-based shRNA approach.22 The selected shRNA-targeted cDNA sequence for GCL catalytic subunit was 5′-GAAGGAGGCTACTTCTATA-3′ (corresponding to nt 772-790; accession number NM_001498).23 To knockdown PARP-1 we used a recently reported shRNA-targeted rat cDNA sequence (5′-GGAGTTGCTCATCTTCAAC-3′, corresponding to nt 789-807; accession number NM_013063).14 As controls, we used the firefly luciferase-targeted oligonucleotide 5′-CTGACGCGGAATACTTCGA-3′, as reported previously.34 All sequences were BLAST-confirmed for specificity. The forward and reverse synthetic 64-nt oligonucleotides (Isogen Life Technologies, Maarsen, The Netherlands) were designed, annealed, and inserted into the BglII/HindIII sites of the pSuper-neo/gfp vector, following the manufacturer's instructions (Oligoengine, Seatle, WA). These constructs express 19 base-pair 9-nt stem-loop shRNAs targeted against the GCL catalytic subunit, PARP-1 or luciferase (control shRNA) mRNAs. The concomitant expression of green fluorescent protein (GFP) from these vectors allowed the identification of transfected cells by fluorescence microscopy and flow cytometry. For the experiments involving PARP-1 overexpression, we used a pEXPRESS mammalian expression vector (4.12 kb) encoding the full-length PARP-1 rat cDNA (3.09 kb; accession number BC085765) (pEXPRESS-PARP-1). This plasmid was purchased from, and sequence-verified by Geneservice Lltd. (cDNA clone number MGC:93658, IMAGE:7107130; Cambridge, UK). To identify transfected cells by flow cytometry, pEXPRESS-PARP-1 and the GFP-encoding empty vector pd2EGFP-N1 (ClonTech Laboratories, Palo Alto, CA, USA) were used in the ratio 3 : 1.
Neurons in primary culture
Cerebral cortex neurons in primary culture were prepared from foetal Wistar rats at 16 days of gestation.35 Dissociated cell suspensions were plated at a density of 2.5 × 105 cells/cm2 in six- or 12-well plates previously coated with poly-D-lysine (15 μg/ml) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal calf serum (FCS). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2/95% air. Forty-eight hours after plating, the medium was replaced with DMEM supplemented with 5% horse serum, 20 mM D-glucose and cytosine arabinoside (10 μM) to prevent non-neuronal proliferation. Neurons were transfected on day 6, when approximately 99% of the cells were Map2-positive.
HEK293T cells
HEK293T cells were used in this study only for confirming the effectiveness and specificity of the GCL shRNA construct, because these cells can be transfected with high efficiency. Cells were maintained in DMEM supplemented with 10% (v/v) FCS and re-seeded at 105 cells/cm2 1 day before transfection, after which FCS was reduced to 0.5% (v/v).
Cell transfections
These were performed with pSuper-neo/gfp-derived plasmid constructions using Lipofectamine 2000 (Invitrogen, Groningen, The Netherlands), following the manufacturer's instructions. Primary neurons were transfected in fresh medium after 6 days in vitro, that is, when cells displayed terminally differentiated neuronal phenotype. After 6 h, the medium was removed and cells were further incubated for 6, 16, 24 or 48 h in DMEM before the immunocytochemical and flow cytometric analyses. The transfection efficiencies were approximately 7%.
Site-directed mutagenesis
To assess whether GCL shRNA was specific, wild-type (GCL) or mutant (GCLmut) forms of catalytic subunit of rat GCL full-length cDNA were used. GCLmut was obtained by PCR site-directed mutagenesis, using pIRES-GCL catalytic subunit23 as a template, followed by DpnI digestion (QuikChange XL, Stratagene). The selected PCR forward and reverse 50-nt oligonucleotides carried silent third codon-base point mutations within the rat cDNA-corresponding GCL shRNA-target sequence (5′-GAAAGAAGCCACGTCAGTT-3′; mutated nucleotides underlined).
Antibodies against GCL
Polyclonal antibodies against the catalytic subunit of rat GCL were obtained by rabbit immunization with the synthetic peptide (Mimotopes Pty. Ltd., Clayton Victoria, Australia) 16QRHADHVRRHGILQ29-C. Rabbits were immunized with 100 ng of the peptides coupled to the haemocyanin from Keyhole Limpet. In order to improve the detection of the proteins, antibodies were purified from the antisera by affinity chromatography using columns of cyanogen-bromide-activated sepharose coupled to the peptides.
Western blotting
Aliquots of whole-cell lysates were subjected to sodium docedyl sulfate-polyacrylamide gel electrophoresis on a 10% acrylamide gel (MiniProtean®, Bio-Rad). After blocking, membranes were immunoblotted against anti-GCL, anti-PAR (Trevigen, Gaithersburg, MD, USA) or anti-Tubulin (Tub; Sigma), which was used as loading control. After incubation with horseradish peroxidase-conjugated goat antirabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), membranes were developed and exposed.
Glutathione concentrations
Cells were washed with ice-cold PBS and immediately scraped off the plastic with 1% (wt/vol) sulfosalicylic acid. Cell lysates were centrifuged at 13 000 × g for 5 min at 4°C and the supernatants were used for the determination of total glutathione (reduced glutathione +2 × oxidized glutathione) and oxidized glutathione on the same day as described previously.36
Flow cytometric analysis of mitochondrial membrane potential
Following treatment, the culture medium was removed and neurons were detached from the plates using 1 mM EDTA (tetrasodium salt) in PBS (pH 7.4). After 1 min at room temperature, cells were carefully re-suspended using a micropipette with a 1-ml tip. MitoProbe™ DiC1(5) Assay Kit for Flow Cytometry (Molecular Probes Europe BV, Leiden, Netherlands) was used to determine quantitatively the Δψm in detached cells by flow cytometry. After treatment, neurons were stained with MitoProbe™ following the manufacturer's instructions, and the GFP+-stained cells were analysed on the FL1 and FL4 channels of a FACScalibur flow cytometer (15 mW argon ion laser tuned at 488 nm; CellQuest software, Becton-Dickinson Biosciences, San José, CA, USA) and expressed as percentages. We used the mitochondrial uncoupler carbonyl cyanide p-[trifluoromethoxy]phenyl hydrazone (FCCP, 5 μM; 5 min) to define the 0% Δψm values; values obtained in control shRNA-treated cells were considered to be 100%. Preliminary experiments carried out to validate this protocol indicated that the values obtained after adding the mitochondrial uncoupler FCCP were similar to those obtained with antimycin A (10 μg/ml) (not shown).
Flow cytometric analysis of apoptotic cell death
Following treatment, cells were carefully detached from the plates using 1 mM EDTA (tetrasodium salt) in PBS (pH 7.4). APC-conjugated annexin-V and 7-AAD (Becton Dickinson Biosciences) were used to determine quantitatively the percentage of apoptotic cells. Cells were stained with annexin V-APC and 7-AAD, following the manufacturer's instructions, and were analysed on a flow cytometer as above. The analyzer threshold was adjusted on the flow cytometer channel to exclude most of the subcellular debris in order to reduce the background noise owing to the neurite disruption during neuronal detaching. To quantify neurons displaying phosphatidylserine translocation on the cell soma plasma membrane, we determined those annexin V-APC-positive cells that were 7-AAD negative. As 7-AAD only binds DNA to those cells having a non-intact plasma membrane (due both to disruption during detaching and necrosis by the treatments), GFP+ annexin V-APC-stained cells that were 7-AAD-negative were considered apoptotic.35, 37
Determination of apoptotic cell death by TUNEL assay
To assess apoptotic death in non-detached, intact neurons, neuronal apoptosis was also quantified by TUNEL assay (Roche Diagnostics, Heidelberg, Germany) following the manufacturer's instructions. Each data point was obtained by counting 500 cells from 10 microscopic fields.
Flow cytometric detection of active caspase-3 and cleaved PARP-1
Active caspase-3 was detected using the APO ACTIVE 3™ Kit (Bachem, San Carlos, CA, USA), following the manufacturer's instructions. Flow cytometric detection of cleaved PARP-1 was achieved using the 3-phycoerythrine conjugate (PE) F21-852 monoclonal antibody (PE mouse anti-cleaved PARP-1; Becton Dickinson Biosciences). This antibody specifically reacts with the 89-kDa PARP-1 domain that is downstream of the caspase-3 cleavage site. Following the manufacturer's instructions, the positive controls were performed by incubation of the cells with etoposide (1 μM) for 24 h before PE mouse anti-cleaved PARP-1 staining.
Flow cytometric analysis of BrdU incorporation into DNA
This was achieved after 18 h of incubation with 10 μg/ml of BrdU using the APC BrdU Flow Kit (Becton Dickinson Biosciences), following the manufacturer's instructions.
Immunocytochemistry
This. was carried out on neurons grown on glass coverslips, which were fixed for 30 min in PBS containing 4% paraformaldehyde, rinsed with PBS and permeabilized for 5 min with 0.3% Triton X-100. Cells were then incubated for 30 min at room temperature in PBS containing blocking solution (0.1% triton X-100, 5% horse serum) and overnight at 4°C with anti-Map2 (Sigma, 1 : 200 dilution), anti-GCL (1 : 40 dilution), anti-PARP-1 (Cell Signalling Technology, 1 : 80 dilution), anti-PAR (Trevigen, 1 : 50 dilution) or anti-AIF (Cell Signalling Technology, 1 : 100 dilution) in the same solution. After washing with PBS, cells were incubated in PBS containing the secondary antibody (anti-mouse-IgG-Cy3, Sigma, 1 : 400 dilution) for 1 h at room temperature. Coverslips were washed and mounted in glycerol/PBS (90/10, by vol) on glass slides for phase-contrast or fluorescence microphotographs at × 10, × 20, or × 60 magnification.
Oxygen radical detection
Cells were incubated in PBS containing 1 μg/ml of dihydrorhodamine-123 (Sigma) at 37°C in the dark for 1 h. Excess dihydrorhodamine-123 was removed by extensive washing with PBS, and the oxidized product, Rh-123, was detected by fluorescence (510–560 nm) microscopy. In addition, we used dihydroethidine (HE; Molecular Probes, Eugene, OR, USA) to confirm oxygen radical formation by the neurons.38 After transfections, cells were incubated in DMEM either in the absence or in the presence of tempol (100 μM) for 24 h. Cells were then incubated in PBS containing 10 μM HE at 37°C in the dark for 45 min. Excess HE was removed by washing with PBS, and the oxidized product (ethidium) was detected by fluorescence (510–560 nm) microscopy. To achieve maximum fluorescence intensity, cells were incubated in the HE solution containing H2O2 (500 μM).
Statistical analysis
Measurements from individual cultures were always performed in triplicate, and the results were expressed as mean±S.E.M. values across three different culture preparations (n=3). Statistical analysis of the results was performed by one-way analysis of variance, followed by the least significant difference multiple range test. In all cases, P<0.05 was considered significant.
Abbreviations
- 7-AAD:
-
7-amino-actinomycin D
- ABA:
-
3-aminobenzamide (ABA)
- AIF:
-
apoptotis-inducing factor
- AIQ:
-
5-aminoisoquinolinone
- BAPTA, 1,2-bis(o-aminophenoxy)ethane-N,N,N′N′-tetraacetic acid tetra(acetoxymethyl) ester∣BrdU:
-
bromodeoxyuridine
- CaMK-II:
-
calcium-calmodulin kinase-II
- DMEM:
-
Dulbecco's modified Eagle's medium
- Δψm:
-
mitochondrial membrane potential
- FCS:
-
foetal calf serum
- GCL:
-
glutamate-cysteine ligase
- GFP:
-
green fluorescent protein
- GSH:
-
reduced glutathione
- GSSG:
-
oxidized glutathione
- GSx:
-
total glutathione (GSH+2 × GSSG)
- HE:
-
hydroethidine
- HEK293T:
-
human embryonic kidney 293T
- KIF4:
-
kinesin superfamily protein 4
- L-BSO:
-
L-buthionine sulfoximine (L-BSO)
- PAR:
-
poly (ADP-ribose) polymers
- PARP-1:
-
poly(ADP-ribose) polymerase-1
- Rh-123:
-
rhodamine-123
- RNAi:
-
RNA interference
- shRNA:
-
small hairpin RNA
References
Ratan RR, Murphy TH, Baraban JM . Oxidative stress induces apoptosis in embryonic cortical neurons. J Neurochem 1994; 62: 376–379.
Bolaños JP, Heales SJR, Land JM, Clark JB . Effect of peroxynitrite on the mitochondrial respiratory chain: differential susceptibility of neurones and astrocytes in primary cultures. J Neurochem 1995; 64: 1965–1972.
Hirrlinger J, Resch A, Gutterer JM, Dringen R . Oligodendroglial cells in culture effectively dispose of exogenous hydrogen peroxide: comparison with cultured neurones, astroglial and microglial cells. J Neurochem 2002; 82: 635–844.
Boyd-Kimball D, Sultana R, Abdul HM, Butterfield DA . Gamma-glutamylcysteine ethyl ester-induced up-regulation of glutathione protects neurons against Abeta(1-42)-mediated oxidative stress and neurotoxicity: implications for Alzheimer's disease. J Neurosci Res 2005; 79: 700–706.
Klein JA, Ackerman SL . Oxidative stress, cell cycle, and neurodegeneration. J Clin Invest 2003; 111: 785–793.
Andersen JK . Oxidative stress in neurodegeneration: cause or consequence? Nat Med 2004; 10 (Suppl): S18–S25.
Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J et al. Gene regulation and DNA damage in the ageing human brain. Nature 2004; 429: 883–891.
Lindahl T, Satoh MS, Poirier GG, Klungland A . Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci 1995; 20: 405–411.
Huber A, Bai P, de Murcia JM, de Murcia G . PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amsterdam) 2004; 3: 1103–1118.
Kim MY, Zhang T, Kraus WL . Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 2005; 19: 1951–1967.
Zhang J, Dawson VL, Dawson TM, Snyder SH . Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science 1994; 263: 687–689.
Szabó C, Oshima H . DNA damage induced by peroxynitrite: subsequent biological effects. Nitric Oxide 1997; 1: 373–385.
Tapodi A, Debreceni B, Hanto K, Bognar Z, Wittmann I, Gallyas FJ et al. Pivotal role of Akt activation in mitochondrial protection and cell survival by poly(ADP-ribose)polymerase-1 inhibition in oxidative stress. J Biol Chem 2005; 280: 35767–35777.
Tanaka S, Takehashi M, Iida S, Kitajima T, Kamanaka Y, Stedeford T et al. Mitochondrial impairment induced by poly(ADP-ribose) polymerase-1 activation in cortical neurons after oxygen and glucose deprivation. J Neurochem 2005; 95: 179–190.
Koh DW, Dawson TM, Dawson VL . Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res 2005; 52: 5–14.
Alano CC, Swanson RA . Players in the PARP-1 cell-death pathway: JNK1 joins the cast. Trends Biochem Sci 2006; 31: 309–311.
Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA 2006; 103: 18308–18313.
Merlo D, Di Stasi AM, Bonini P, Mollinari C, Cardinale A, Cozzolino F et al. DNA repair in post-mitotic neurons: a gene-trapping strategy. Cell Death Differ 2005; 12: 307–309.
Griffith OW . Mechanism of action, metabolism, and toxicity of buthionine sulfoximine and its higher homologs, potent inhibitors of glutathione synthesis. J Biol Chem 1982; 257: 13704–13712.
Orlowski M, Meister A . Isolation of highly purified γ-glutamylcysteine synthetase from rat kidney. Biochemistry 1971; 10: 372–380.
Ikeda Y, Hokamura K, Kawai T, Ishiyama J, Ishikawa K, Anraku T et al. Neuroprotective effects of KCL-440, a new poly(ADP-ribose) polymerase inhibitor, in the rat middle cerebral artery occlusion model. Brain Res 2005; 1060: 73–80.
Brummelkamp TR, Bernards R, Agami R . A system for stable expression of short interfering RNAs in mammalian cells. Science 2002; 296: 550–553.
Diaz-Hernandez JI, Almeida A, Delgado-Esteban M, Fernandez E, Bolanos JP . Knockdown of glutamate-cysteine ligase by small hairpin RNA reveals that both catalytic and modulatory subunits are essential for the survival of primary neurons. J Biol Chem 2005; 280: 38992–39001.
Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM . Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem 1998; 273: 33533–33539.
Yu SW, Wang H, Poitras M, Coombs C, Bowers WJ, Federoff HJ et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 2002; 297: 259–263.
Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W et al. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci 2004; 24: 10963–10973.
Xu Y, Huang S, Liu ZG, Han J . Poly(ADP-ribose) polymerase-1 signaling to mitochondria in necrotic cell death requires RIP1/TRAF2-mediated JNK1 activation. J Biol Chem 2006; 281: 8788–8795.
Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N et al. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J 2000; 14: 729–739.
Midorikawa R, Takei Y, Hirokawa N . KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell 2006; 125: 371–383.
Critchlow SE, Jackson SP . DNA end-joining: from yeast to man. Trends Biochem Sci 1998; 23: 394–398.
Virag L, Salzman AL, Szabo C . Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. J Immunol 1998; 161: 3753–3759.
Riedl SJ, Shi Y . Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 2004; 5: 897–906.
Frost MT, Wang Q, Moncada S, Singer M . Hypoxia accelerates nitric oxide-dependent inhibition of mitochondrial complex I in activated macrophages. Am J Physiol Regul Integr Comp Physiol 2005; 288: R394–400.
Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI et al. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat Cell Biol 2004; 6: 121–128.
Almeida A, Moncada S, Bolaños JP . Nitric oxide switches on glycolysis through the AMP protein kinase and 6-phosphofructo-2-kinase pathway. Nat Cell Biol 2004; 6: 45–51.
Garcia Nogales P, Almeida A, Bolaños JP . Peroxynitrite protects neurons against nitric oxide-mediated apoptosis. A key role for glucose-6-phosphate dehydrogenase activity in neuroprotection. J Biol Chem 2003; 278: 864–874.
Almeida A, Bolanos JP, Moreno S . Cdh1/Hct1-APC is essential for the survival of postmitotic neurons. J Neurosci 2005; 25: 8115–8121.
Bindokas VP, Jordan J, Lee CC, Miller RJ . Superoxide production in rat hippocampal neurons: selective imaging with hydroethidine. J Neurosci 1996; 16: 1324–1336.
Acknowledgements
This work was supported by the Fondo de Investigación Sanitaria (Grant FIS06/0794 to AA), Junta de Castilla y León (Grants SA082A05 to AA; SA081/04 to JPB), Ministerio de Educación y Ciencia (Grant SAF2004-2038 to JPB) and an MRC Programme Grant to SM. AA is funded by the Consejería de Sanidad de Castilla y León and Instituto de Salud Carlos III. We wish to thank Ms. Mónica Resch for technical assistance and Annie Higgs for help in the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by L Greene
Rights and permissions
About this article
Cite this article
Diaz-Hernandez, J., Moncada, S., Bolaños, J. et al. Poly(ADP-ribose) polymerase-1 protects neurons against apoptosis induced by oxidative stress. Cell Death Differ 14, 1211–1221 (2007). https://doi.org/10.1038/sj.cdd.4402117
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402117
Keywords
This article is cited by
-
The Extract of Leonurus sibiricus Transgenic Roots with AtPAP1 Transcriptional Factor Induces Apoptosis via DNA Damage and Down Regulation of Selected Epigenetic Factors in Human Cancer Cells
Neurochemical Research (2018)
-
Nicotinamide Inhibits Ethanol-Induced Caspase-3 and PARP-1 Over-activation and Subsequent Neurodegeneration in the Developing Mouse Cerebellum
The Cerebellum (2018)
-
γ-Glutamylcysteine detoxifies reactive oxygen species by acting as glutathione peroxidase-1 cofactor
Nature Communications (2012)
-
Induction of apoptosis by the inhibitors of poly(ADP-ribose)polymerase in HeLa cells
Molecular and Cellular Biochemistry (2009)
-
Neuroprotective Role of Antidiabetic Drug Metformin Against Apoptotic Cell Death in Primary Cortical Neurons
Journal of Molecular Neuroscience (2008)