Abstract
Apoptotic cell death in the nematode C. elegans culminates with the removal of the dying cells from the organism. This removal is brought forth through a rapid and specific engulfment of the doomed cell by one of its neighbors. Over half a dozen genes have been identified that function in this process in the worm. Many of these engulfment genes have functional homologs in Drosophila and higher vertebrates. Indeed, there is growing evidence supporting the hypothesis that the pathways that mediate the removal of apoptotic cells might be, at least in part, conserved through evolution. Cell Death and Differentiation (2001) 8, 564–568
Similar content being viewed by others
Introduction
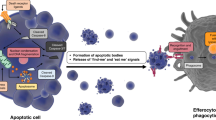
It seems ironic that we should be asking trade secrets of that greatest of undertakers, the worm. Much understanding of the removal of cellular corpses has come from studies of professional macrophages and immune cells. Given the caveat that the nematode Caenorhabditis elegans has neither professional macrophages nor an immune system, one could still argue that the genetic conservation and tractability of the worm–and its ability to survive a loss of engulfment potential–provide a sound basis for its use as a model organism for phagocytosis. Indeed, research on the worm has already shed some light on a process that is riddled with conceptual black boxes (see Figure 1). This process starts in the doomed cell, with the molecular execution pathway triggering or activating ‘eat me’ signals that need to be presented at the plasma membrane for recognition by the engulfing cell. The engulfing cell must be able to receive and interpret these signals, relay them to the cytoskeleton, and then bind to and extend cytoplasmic arms around the dying cell. Lastly, the engulfing cell must recycle the remains of its prey.
The nematode C. elegans has already proved instrumental in studying fundamental questions in developmental biology. Its benefits as a model organism include its powerful genetics,1 a fully sequenced genome,2 and its simple, reproducible pattern of cell divisions during development.3,4,5
During this development, a large number of cells undergo programmed cell death (PCD, apoptosis). In hermaphrodites, 131 of the 1090 cells generated die, while in males 147 of the 1178 cells born die.3,4,5 The identity, time and location of each of these deaths is highly reproducible, giving researchers the ability to study PCD and engulfment in vivo with single-cell resolution. Apoptotic cell death is also prevalent in the adult hermaphrodite, where almost half of all germ cells undergo programmed cell death.6
Apoptotic cells are engulfed and degraded soon after they are generated, a process that normally takes less than an hour.7 Since C. elegans has no professional phagocytic cells, the task of removing dying cells from the body falls to non-professional neighbors surrounding the dying cell.5,7 For example, dying germ cells are removed by the myoepithelial somatic sheath cells that surround the gonad.6
Genetic studies in C. elegans have resulted in the isolation of several mutations that affect engulfment (see Table 1 and below).8,9,10 These mutations identify seven genes that function in two pathways for phagocytosis. The first group is defined by members of the Rac pathway: ced-2, ced-5, and ced-10 encode nematode homologs of Crkll, DOCK180, and Rac1, respectively.11,12 A fourth gene, ced-12, also acts in this pathway but its molecular identity has not yet been reported.10 A second group that affects engulfment in the nematode is composed of membrane proteins CED-1 and CED-7 and an adaptor protein, CED-6. ced-1 encodes a protein with sequence and structural similarity to the scavenger receptor SREC.13 ced-7 is an ABC1 transporter homolog.14 ced-6 contains a phosphotyrosine binding (PTB) domain,15 a motif that is found in some adaptor proteins like Shc and IRS-1.16
From executioners to undertakers
In C. elegans and other species, there exists a gap in our understanding between caspase activation and presentation of ‘eat me’ signals at the external plasma membrane. C. elegans has four known caspases, with CED-3 being the most extensively studied.17 Several candidate CED-3 substrates have been reported, notably itself and CSP-2B as proproteins, CED-9, the viral caspase inhibitor p35, and FEM-3, a protein involved in regulating sex determination.17,18,19,20 However, how substrate cleavage mediates events downstream of the execution step is currently unknown in worms. In fact, presentation of ‘eat me’ signals may not depend entirely on caspases: nematodes that have an inactivated CED-3 still have very low levels of cell death, and these cell corpses linger longer if an engulfment gene is also mutated6,21 (D Hoeppner and MO Hengartner, unpublished). Furthermore, the same engulfment pathway is also used to remove cells that die via CED-3-independent mechanisms, such as the necrotic-like deaths observed following hyperactivation of various types of ion channels.10 Cells that die by a variety of insults, like constitutively activated ion channels or G proteins, linger longer in the body if any of the engulfment genes is inactivated. Even cells that are only ‘sick’ have a greater chance of survival if the engulfment machinery is sabotaged, as demonstrated by Kim and Horvitz in their study of the genes lin-24 and lin-33. Rare gain-of-function mutations in these genes lead to the inappropriate death of the Pn.p cells, a set of larval blast cells required for vulval development.22 The mechanism underlying these atypical deaths is unclear, but appears to be independent of the classical apoptotic pathway,23 as the deaths are not suppressed in, e.g., ced-3 mutants.22 In contrast, mutations in at least some engulfment genes partially rescue the Pn.p cells from death.23 That blocking engulfment rescues from death suggests that in these mutants, the Pn.p cells are often not dead but simply sick, but that an early activation of the engulfment pathway results in them being ‘eaten alive.’ Preventing engulfment might give the cells a chance to recover and function properly. A corollary of this hypothesis is that the Pn.p cells express ‘eat me’ signals well before they are dead. Early recognition of doomed cells might in fact be a general phenomenon in C. elegans. An extreme example of this idea has been reported by Robertson and Thomson, whose electron micrographs of a cell fated to die showed that it had already been recognized by an engulfing neighbor cell before it had even finished separating from its mother cell.7
Finally, even a healthy cell can be ‘murdered’ by phagocytosis in C. elegans.24 The linker cell is required to lead the migration of the male gonad to the tail, where the reproductive and digestive system fuse.4 The linker cell dies once it reaches its destination and is engulfed by one of two cells in the proctodeum. This death is required to open a channel between the vas deferens and the cloaca. The linker cell survives if it never reaches the engulfing cells or if the engulfing cells are eliminated by laser microsurgery of their common progenitor cell.24 Interestingly, the linker cell still dies about half of the time in ced-3 mutants, suggesting that the cell death machinery acts in, but is not essential for, the programmed death of the linker cell.25
Worm ‘eat me’ signals
Little is known about ‘eat me’ signals in worms. None of the surface antigens that are presented on the surface of many dying mammalian cells, such as phosphatidylserine (PS), changes in carbohydrate chains on surface proteins and lipids, and ICAM-3 (intercellular adhesion molecule-3), have yet been shown to be recognized by engulfing cells in C. elegans.26 Nonetheless, given the high level of conservation in the process, it would be surprising if these surface antigens were not also major contributing factors in C. elegans engulfment. Interestingly, the CED-7 homolog ABC1 promotes the exposure of phosphatidylserine in mammalian cells.27,28 Furthermore, Fadok and colleagues have found a PS receptor (PSR) that recognizes PS on the surface of apoptotic macrophages; interestingly, the C. elegans genome contains a good potential ortholog of this gene.29 These findings suggest that PS may also be exposed in dying nematode cells.
Recognition of dying cells
A large number of candidate apoptotic receptors have been described in mammals.26 Only a few of these genes have known nematode homologs, though. As mentioned above, C. elegans contains a homolog of PSR; whether this gene is involved in recognition of apoptotic cells remains to be proven. Recently Zhou and Horvitz have shown that ced-1 encodes a transmembrane molecule with some similarity to SREC (scavenger receptor expressed by endothelial cells).13,30 SREC may mediate the endocytosis of chemically modified lipoproteins or phopholipids, and so PS or modified LDL is a contender for CED-1’s currently unknown ligand. CED-7 is also required in engulfing cells.14 Although its purpose in these cells is presently speculative, it is worth noting that its mammalian homolog ABC1 also acts in macrophages, where it participates in lipid transport in the cell membrane.31,32
Necrotic-like cell deaths can also occur in C. elegans through a variety of cellular insults.33 Interestingly, Chung and colleagues recently showed that cells undergoing necrotic-like deaths require the same engulfment genes for removal as cells that undergo programmed cell death.10 A similar situation might exist in mammals; ABC1, the CED-7 homolog, acts in macrophages to remove necrotic cells as well as programmed cell deaths.34 Other studies, however, have suggested that the receptors recognizing apoptotic cells and necrotic cells are distinct.35 Clearly, more work on this issue is warranted.
Messengers from the membrane to the cytoskeleton
CED-1 contains several potential tyrosine kinase phosphorylation sites, which in many growth factor receptors mediate interactions with cytoplasmic adaptor proteins that contain SH2 or phosphotyrosine-binding (PTB) domain-containing proteins. CED-6, a PTB domain-containing protein, is one such candidate adaptor protein which might bind CED-1.15 Consistent with this hypothesis, CED-6 acts downstream of CED-1 and CED-7 by genetic analysis. The human CED-6 homolog is also required for phagocytosis. Dimerization of mammalian CED-6 is required for its activity; this dimerization might be important for interaction with its yet unknown partners.36,37,38
Moving membranes
Once a neighboring cell has received a signal from its neighbor, it needs the means to remove the dying cell. This is accomplished by extending cytoplasmic arms around the dying cell. ced-2(Crkll), ced-5(DOCK180), and ced-10(Rac1) have been shown by homology to provide the means for acting on this signal by allowing the engulfing cell to extend processes around the dying cell (Figure 1).11,12 A fourth gene, ced-12, also acts in this cascade in C. elegans, but the molecular nature of this gene has not yet been reported.10 Recruitment of Rac1 to the cytoplasmic membrane has been shown to be involved in pseudopod extension during phagocytosis.39,40,41 In other systems, αvβ5 integrin has been shown to recruit p130Cas to the membrane, which becomes anchored to the membrane upon phosphorylation and subsequently binds to Crkll.42 This complex then recruits DOCK180 and activates Rac1, leading to actin/myosin assembly-based pseudopod extension.43 C. elegans integrins have no reported role in phagocytosis, but do affect the adhesion and migration of many cells.44 A p130Cas homolog has not been reported but may exist in C. elegans (K Ravichandran and W Van Criekinge, personal communication).
Relationship of engulfment to degradation
Nucleases are known to be required for the degradation of DNA in dying cells. Studies by Wu and colleagues have indicated that there are at least three different nucleases involved in this process in C. elegans.45 The first endonuclease is known only by its activity, which results in the creation of TUNEL-reactive DNA. It is dependent on the interaction of the engulfing cell with the dying cell and requires the function of CED-1 and CED-7. The second nuclease, NUC-1/DNase II, further degrades DNA so the DNA is no longer recognized by TUNEL staining. The last nuclease acts only after the dying cell is fully enclosed by the phagocytic cell, suggesting that the engulfing cell may supply this nuclease, perhaps in lysosomes, for the final recycling of the dead cell's DNA.
The roles of caspases, other proteases, and lipases in C. elegans in providing engulfment signals and in degrading the cell provides another black box waiting to be opened. Of interest will be the timing of activation of ‘eat me’ signals in relation to general cell destruction events, and what contributions the dying and engulfing cell each make in removing potentially harmful cells from the body.
Future directions
The powerful genetics of C. elegans has revealed the involvement in phagocytosis of two cell surface proteins (CED-1 and CED-7), a previously uncharacterized adaptor molecule (CED-6), and confirmed the role of the Crkll/DOCK180/Rac pathway (CED-2, CED-5, and CED-10, respectively). Biochemical studies of these molecules, coupled with continued genetic analysis, will continue to further our understanding of cell corpse removal in both the worm and man (see Figure 1).
Abbreviations
- SREC:
-
scavenger receptor of endothelial cells
- PCD:
-
programmed cell death
- PTB:
-
phosphotyrosine binding
- PS:
-
phosphatidylserine
- ICAM-3:
-
intercellular adhesion molecule-3
- PSR:
-
phosphatidylserine receptor
References
Brenner S . 1974 The genetics of Caenorhabditis elegans Genetics 77: 71–94
The C. elegans Sequencing Consortium 1998 Genome Sequence of the nematode C. elegans: a platform for investigating biology Science 282: 2012–2018
Sulston JE, Horvitz HR . 1977 Post-embryonic cell lineages of the nematode, Caenorhabditis elegans Dev. Biol. 56: 110–156
Kimble J, Hirsh D . 1979 The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans Dev. Biol. 70: 396–417
Sulston JE, Schierenberg E, White JG, Thomson JN . 1983 The embryonic cell lineage of the nematode Caenorhabditis elegans Dev. Biol. 100: 64–119
Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO . 1999 Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline Development 126: 1011–1022
Robertson A, Thomson N . 1982 Morphology of programmed cell death in the ventral nerve cord of Caenorhabditis elegans larvae J. Embryol. exp. Morph. 67: 89–100
Hedgecock EM, Sulston JE, Thomson JN . 1983 Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans Science 220: 1277–1279
Ellis RE, Jacobson DM, Horvitz HR . 1991 Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans Genetics 129: 79–94
Chung S, Gumienny TL, Hengartner MO, Driscoll M . 2000 A common set of genes mediate removal of both apoptotic and necrotic cell corpses in C. elegans Nat. Cell Biol. 2: 931–937
Wu Y-C, Horvitz HR . 1998 C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180 Nature 392: 501–504
Reddien PW, Horvitz HR . 2000 CED-2/Crkll and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans Nat. Cell Biol. 2: 131–136
Zhou Z, Hartwieg E, Horvitz HR . 2001 CED-1 is a transmembrane receptor that mediates cell-corpse engulfment in C. elegans Cell In Press
Wu Y-C, Horvitz HR . 1998 The C. elegans cell-corpse engulfment gene ced-7 encodes a protein similar to ABC transporters Cell 93: 951–960
Liu QA, Hengartner MO . 1998 Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans Cell 93: 961–972
Yenush L, White MF . 1997 The IRS-signalling system during insulin and cytokine action Bioessays 19: 491–500
Shaham S . 1998 Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades J. Biol. Chem. 273: 35109–35117
Xue D, Horvitz HR . 1995 Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein Nature 377: 248–251
Yang X, Chang HY, Baltimore D . 1998 Essential role of CED-4 oligomerization in CED-3 activation and apoptosis Science 281: 1355–1357
Chan SL, Yee KS, Tan KM, Yu VC . 2000 The Caenorhabditis elegans sex determination protein FEM-1 is a CED-3 substrate that associates with CED-4 and mediates apoptosis in mammalian cells J. Biol. Chem. 275: 17925–17928
Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR . 1993 The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1beta converting enzyme Cell 75: 641–652
Ferguson EL, Horvitz HR . 1985 Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans Genetics 110: 17–72
Kim S . 1994 Two C. elegans genes that can mutate to cause degenerative cell death. [Ph.D.] Cambridge, MA, USA: MIT
Sulston JE, Albertson DG, Thomson JN . 1980 The Caenorhabditis elegans male: postembryonic development of nongonadal structures Dev. Biol. 78: 542–576
Ellis HM, Horvitz HR . 1986 Genetic control of programmed cell death in the nematode C. elegans Cell 44: 817–829
Savill J, Fadok V . 2000 Corpse clearance defines the meaning of cell death Nature 407: 784–788
Marguet D, Luciani MF, Moynault A, Williamson P, Chimini G . 1999 Engulfment of apoptotic cells involves the redistribution of membrane phosphatidylserine on phagocyte and prey Nat. Cell Biol. 1: 454–456
Hamon Y, Broccardo C, Chambenoit O, Luciani MF, Toti F, Chaslin S, Freyssinet JM, Devaux PF, McNeish J, Marguet D, Chimini G . 2000 ABC1 promotes engulfment of apoptotic cells and transbilayer redistribution of phosphatidylserine Nat. Cell Biol. 2: 399–406
Fadok VA, Bratton DL, Rose DM, Pearson A, Ezekewitz RA, Henson PM . 2000 A receptor for phosphatidylserine-specific clearance of apoptotic cells Nature 405: 85–90
Adachi H, Tsujimoto M, Arai H, Inoue K . 1997 Expression cloning of a novel scavenger receptor from human endothelial cells J. Biol. Chem. 272: 31217–31220
Borst P, Zelcer N, van Helvoort A . 2000 ABC transporters in lipid transport Biochim. Biophys. Acta. 1486: 128–144
Hamon Y, Luciani MF, Becq F, Verrier B, Rubartelli A, Chimini G . 1997 Interleukin-1beta secretion is impaired by inhibitors of the ATP binding cassette transporter, ABC1 Blood 90: 2911–2915
Hall DH, Gu G, Garcia-Anoveros J, Gong L, Chalfie M, Driscoll M . 1997 Neuropathology of degenerative cell death in Caenorhabditis elegans J. Neurosci. 17: 1033–1045
Moynault A, Luciani MF, Chimini G . 1998 ABC1, the mammalian homologue of the engulfment gene ced-7, is required during phagocytosis of both necrotic and apoptotic cells Biochem. Soc. Trans 26: 629–635
Hirt UA, Gantner F, Leist M . 2000 Phagocytosis of nonapoptotic cells dying by caspase-independent mechanisms J. Immunol. 164: 6520–6529
Liu QA, Hengartner MO . 1999 Human CED-6 encodes a functional homologue of the Caenorhabditis elegans engulfment protein CED-6 Curr. Biol. 9: 1347–1350
Smits E, Van Criekinge W, Plaetinck G, Bogaert T . 1999 The human homologue of Caenorhabditis elegans CED-6 specifically promotes phagocytosis of apoptotic cells Curr. Biol. 9: 1351–1354
Su HP, Brugnera E, Van Criekinge W, Smits E, Hengartner M, Bogaert T, Ravichandran KS . 2000 Identification and characterization of a dimerization domain in CED-6, an adapter protein involved in engulfment of apoptotic cells J. Biol. Chem. 275: 9542–9549
Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S . 1997 Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes J. Exp. Med. 186: 1487–1494
Caron E, Hall A . 1998 Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases Science 282: 1717–1721
Castellano F, Montcourrier P, Chavrier P . 2000 Membrane recruitment of rac1 triggers phagocytosis J. Cell Sci. 113: 2955–2961
Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA . 1998 CAS/Crk coupling serves as a ‘molecular switch’ for induction of cell migration J. Cell Biol. 140: 961–972
Cheresh DA, Leng J, Klemke RL . 1999 Regulation of cell contraction and membrane ruffling by distinct signals in migratory cells J. Cell Biol. 146: 1107–1116
Brown NH . 2000 Cell-cell adhesion via the ECM: integrin genetics in fly and worm Matrix Biol. 19: 191–201
Wu YC, Stanfield GM, Horvitz HR . 2000 NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis Genes Dev. 14: 536–548
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by M Piacentini
Rights and permissions
About this article
Cite this article
Gumienny, T., Hengartner, M. How the worm removes corpses: the nematode C. elegans as a model system to study engulfment. Cell Death Differ 8, 564–568 (2001). https://doi.org/10.1038/sj.cdd.4400850
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4400850
Keywords
This article is cited by
-
Dynein links engulfment and execution of apoptosis via CED-4/Apaf1 in C. elegans
Cell Death & Disease (2018)
-
Your neighbours matter – non-autonomous control of apoptosis in development and disease
Cell Death & Differentiation (2016)
-
Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans
Cell Death & Differentiation (2014)
-
Cell-nonautonomous inhibition of radiation-induced apoptosis by dynein light chain 1 in Caenorhabditis elegans
Cell Death & Disease (2013)
-
The c-Jun kinase signaling cascade promotes glial engulfment activity through activation of draper and phagocytic function
Cell Death & Differentiation (2013)