Abstract
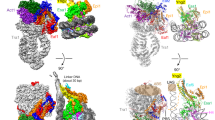
Gene activation is a highly regulated process that requires the coordinated action of proteins to relieve chromatin repression and to promote transcriptional activation. Nuclear histone acetyltransferase (HAT) enzymes provide a mechanistic link between chromatin destabilization and gene activation by acetylating the ε-amino group of specific lysine residues within the amino-terminal tails of core histones to facilitate access to DNA by transcriptional activators1,2. Here we report the high-resolution crystal structure of the HAT domain of Tetrahymena GCN5 (tGCN5) bound with both its physiologically relevant ligands, coenzyme A (CoA) and a histone H3 peptide, and the structures of nascent tGCN5 and a tGCN5/acetyl-CoA complex. Our structural data reveal histone-binding specificity for a random-coil structure containing a G-K-X-P recognition sequence, and show that CoA is essential for reorienting the enzyme for histone binding. Catalysis appears to involve water-mediated proton extraction from the substrate lysine by a glutamic acid general base and a backbone amide that stabilizes the transition-state reaction intermediate. Comparison with related N-acetyltransferases indicates a conserved structural framework for CoA binding and catalysis, and structural variability in regions associated with substrate-specific binding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Grant,P. A., Sterner,D. E., Duggan,L. J., Workman,J. L. & Berger,S. L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8, 193–197 (1998).
Mizzen,C. A. & Allis,C. D. Linking histone acetylation to transcriptional regulation. Cell. Mol. Life Sci. 54, 6–20 (1998).
Trievel,R. C. et al. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl Acad. Sci. USA 96, 8931–8936 (1999).
Clements,A. et al. Crystal structure of the histone acetyltransferase domain of the human P/CAF transcriptional regulator bound to coenzyme-A. EMBO J. 18, 3521–3532 (1999).
Neuwald,A. F. & Landsman,D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that include the yeast SPT10 protein. Trends Biochem. Sci. 22, 154–155 (1997).
Dutnall,R. N., Tafrov,S. T., Sternglanz,R. & Ramakrishnan,V. Structure of the histone acetyltransferase Hat1: A paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 94, 427–438 (1998).
Wolf,E. et al. Crystal structure of a GCN5-related N-acetyltransferase: Serratia maracescens aminoglycoside 3-N-acetyltransferase. Cell 94, 439–449 (1998).
Hickman,A., Namboodiri,M. A. A., Klein,D. C. & Dyda,F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: Enzyme complex at 1.8 Å resolution with a bisubstrate analog. Cell 97, 361–369 (1999).
Hickman,A. B., Klein,D. C. & Dyda,F. Melatonin biosynthesis: The structure of serotonin N-acetyltransferase at 2.5 angstrom resolution suggests a catalytic mechanism. Mol. Cell 3, 23–32 (1999).
Lin,Y., Fletcher,M., Zhou,J., Allis,C. D. & Wagner,G. Solution structure of the catalytic domain of Tetrahymena GCN5 histone acetyltransferase in complex with coenzyme A. Nature 400, 86–89 (1999).
Tanner,K. G. et al. Catalytic mechanism and function of invariant glutamic acid-173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274, 18157–18160 (1999).
Kuo,M. H., Zhou,J. X., Jambeck,P., Churchill,M. E. A. & Allis,C. D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12, 627–639 (1998).
Wang,L., Liu,L. & Berger,S. L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12, 640–653 (1998).
Grant,P. A. et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650 (1997).
Grant,P. A. et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900 (1999).
Weston,S. A. et al. Crystal structure of the anti-fungal target N-myristoyltransferase. Nature Struct. Biol. 5, 213–221 (1998).
Bhatnagar,R. S. et al. Structure of N-myristoyltransferase with bound myristoylCoA and peptide substrate analogs. Nature Struct. Biol. 5, 1091–1097 (1998).
Otwinowski,Z. in Proceedings of the CCP4 Study Weekend: Data collection and processing (eds Sawyer, L., Isaacs, N. & Bailey, S.) 56–62 (SERC Daresbury Laboratory, Warrington, UK, 1993).
Leslie,A. G. W. in CCP4 and ESF-EACMB Newsletter on Protein Crystallography (Daresbury Laboratory, Daresbury, UK, 1992).
Furey,W. & Swaminathan,S. in Methods in Enzymology (eds Carter, C. W. & Sweet, R. M.) 590–620 (Academic, Orlando, 1997).
Jones,T. A., Zou,J. Y. & Cowen,S. W. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger,A. T. X-PLOR Manual, Version 3.8 (Yale Univ. Press, New Haven, 1996).
Brunger,A. T. & Krukowski,A. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. A 46, 585–593 (1990).
Rice,L. M. & Brunger, A T. Torsion angle dynamics: Reduced variable conformational sampling enhances crystallographic structure refinement. Proteins 19, 277–290 (1994).
Jiang,J. S. & Brunger,A. T. Protein hydration observed in X-ray diffraction: solvation properties of penicillopepsin and neuraminidase crystal structures. J. Mol. Biol. 243, 100–115 (1994).
Brunger,A. T., Kuriyan,J. & Karplus,M. Crystallographic R factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Navaza,J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Brunger,A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones,T. A. A graphics model building and refinement system for macromolecules. J. Appl. Crystallogr. 11, 268–272 (1978).
Acknowledgements
We thank J. Berendzen, B. Sweet and their staff for access to and help on beamline X8C at NSLS; D. Thiel and his staff for access to and help on beamlines A1 and F2 at CHESS; and R. Venkataramani, A. Clements, T. Stams and D. King for useful discussions. This work was supported by NIH grants to R.M., S.L.B. and C.D.A., an NIH Under-represented Minority Supplement to J.R.R., a Howard Hughes predoctoral fellowship to R.C.T. and a grant from the Fannie E. Rippel Foundation to R.M.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rojas, J., Trievel, R., Zhou, J. et al. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 (1999). https://doi.org/10.1038/43487
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/43487
This article is cited by
-
Quantitative proteomic analysis of the lysine acetylome reveals diverse SIRT2 substrates
Scientific Reports (2022)
-
Crystal structure of the phage-encoded N-acetyltransferase in complex with acetyl-CoA, revealing a novel dimeric arrangement
Journal of Microbiology (2022)
-
Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins
Scientific Reports (2021)
-
Coordination of microtubule acetylation and the actin cytoskeleton by formins
Cellular and Molecular Life Sciences (2018)
-
CBP/p300: intramolecular and intermolecular regulations
Frontiers in Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.