Abstract
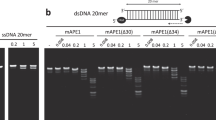
Human apurinic/apyrimidinic endonuclease (APE1) is an essential enzyme in DNA base excision repair that cuts the DNA backbone immediately adjacent to the 5′ side of abasic sites to facilitate repair synthesis by DNA polymerase β (ref. 1). Mice lacking the murine homologue of APE1 die at an early embryonic stage2. Here we report that APE1 has a DNA exonuclease activity on mismatched deoxyribonucleotides at the 3′ termini of nicked or gapped DNA molecules. The efficiency of this activity is inversely proportional to the gap size in DNA. In a base excision repair system reconstituted in vitro, the rejoining of nicked mismatched DNA depended on the presence of APE1, indicating that APE1 may increase the fidelity of base excision repair and may represent a new 3′ mispaired DNA repair mechanism. The exonuclease activity of APE1 can remove the anti-HIV nucleoside analogues 3′-azido-3′-deoxythymidine and 2′,3′-didehydro-2′, 3′-dideoxythymidine from DNA, suggesting that APE1 might have an impact on the therapeutic index of antiviral compounds in this category.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wilson, D. M., Takeshita, M., Grollman, A. P. & Demple, B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 270, 16002–16007 (1995).
Xanthoudakis, S., Smeyne, R. J., Wallace, J. D. & Curran, T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA 93, 8919–8923 (1996).
Lindahl, T. Suppression of spontaneous mutagenesis in human cells by DNA base excision–repair. Mutat. Res. 462, 129–135 (2000).
Loeb, L. A. & Preston, B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 20, 201–230 (1986).
Matsumoto, Y. & Kim, K. Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science 269, 699–702 (1995).
Kunkel, T. A. The mutational specificity of DNA polymerase-β during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J. Biol. Chem. 260, 5787–5796 (1985).
Loeb, K. R. & Loeb, L. A. Significance of multiple mutations in cancer. Carcinogenesis 21, 379–385 (2000).
Demple, B., Herman, T. & Chen, D. S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA 88, 11450–11454 (1991).
Robson, C. N., Milne, A. M., Pappin, D. J. & Hickson, I. D. Isolation of cDNA clones encoding an enzyme from bovine cells that repairs oxidative DNA damage in vitro: homology with bacterial repair enzymes. Nucleic Acids Res. 19, 1087–1092 (1991).
Chou, K. M., Kukhanova, M. & Cheng, Y. C. A novel action of human apurinic/apyrimidinic endonuclease. Excision of l-configuration deoxyribonucleoside analogs from the 3′ termini of DNA. J. Biol. Chem. 275, 31009–31015 (2000).
Grove, K. L. et al. Anticancer activity of β-l-dioxolane-cytidine, a novel nucleoside analogue with the unnatural L configuration. Cancer Res. 55, 3008–3011 (1995).
Wilson, S. H. & Kunkel, T. A. Passing the baton in base excision repair. Nature Struct. Biol. 7, 176–178 (2000).
Bennett, R. A., Wilson, D. M., Wong, D. & Demple, B. Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc. Natl Acad. Sci. USA 94, 7166–7169 (1997).
Husain, I. et al. Purification and characterization of DNA ligase III from bovine testes. Homology with DNA ligase II and vaccinia DNA ligase. J. Biol. Chem. 270, 9683–9690 (1995).
Cheng, Y. C., Gao, W. Y., Chen, C. H., Vazquez-Padua, M. & Starnes, M. C. DNA polymerases versus HIV reverse transcriptase in AIDS therapy. Ann. NY Acad. Sci. 616, 217–223 (1990).
Bouayadi, K. et al. Overexpression of DNA polymerase β sensitizes mammalian cells to 2′,3′-deoxycytidine and 3′-azido-3′-deoxythymidine. Cancer Res. 57, 110–116 (1997).
Skalski, V., Liu, S. H. & Cheng, Y. C. Removal of anti-human immunodeficiency virus 2′,3′-dideoxynucleoside monophosphates from DNA by a novel human cytosolic 3′-5′ exonuclease. Biochem. Pharmacol. 50, 815–821 (1995).
Zhu, Q. Y., Scarborough, A., Polsky, B. & Chou, T. C. Drug combinations and effect parameters of zidovudine, stavudine, and nevirapine in standardized drug-sensitive and resistant HIV type 1 strains. AIDS Res. Hum. Retrovir. 12, 507–517 (1996).
Strauss, P. R., Beard, W. A., Patterson, T. A. & Wilson, S. H. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem. 272, 1302–7. (1997).
McCullough, A. K., Dodson, M. L. & Lloyd, R. S. Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem. 68, 255–285 (1999).
Chen, D. S., Herman, T. & Demple, B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 19, 5907–5914 (1991).
Bhagwat, A. S., Sanderson, R. J. & Lindahl, T. Delayed DNA joining at 3′ mismatches by human DNA ligases. Nucleic Acids Res. 27, 4028–4033 (1999).
Hoss, M. et al. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 18, 3868–3875 (1999).
Mazur, D. J. & Perrino, F. W. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′–5′ exonucleases. J. Biol. Chem. 274, 19655–19660 (1999).
Mazur, D. J. & Perrino, F. W. Excision of 3′ termini by the Trex1 and TREX2 3′-5′ exonucleases: characterization of the recombinant proteins. J. Biol. Chem. 276, 17022–17029 (2001).
Acknowledgements
We thank Z. Hatahet and J. B. Sweasy for discussion; M. Kelley and B. Demple for providing the APE clones; A. Tomkinson for human DNA ligase I; and J. Sweasy for human DNA polymerase β.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Chou, KM., Cheng, YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature 415, 655–659 (2002). https://doi.org/10.1038/415655a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/415655a
This article is cited by
-
Lung cancer risk in workers occupationally exposed to polycyclic aromatic hydrocarbons with emphasis on the role of DNA repair gene
International Archives of Occupational and Environmental Health (2023)
-
APE1 distinguishes DNA substrates in exonucleolytic cleavage by induced space-filling
Nature Communications (2021)
-
Molecular snapshots of APE1 proofreading mismatches and removing DNA damage
Nature Communications (2018)
-
CiAPEX2 and CiP0, candidates of AP endonucleases in Ciona intestinalis, have 3′-5′ exonuclease activity and contribute to protection against oxidative stress
Genes and Environment (2017)
-
Distinct catalytic activity and in vivo roles of the ExoIII and EndoIV AP endonucleases from Sulfolobus islandicus
Extremophiles (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.