Abstract
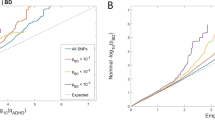
The norepinephrine transporter (NET) gene is an attractive candidate gene for attention-deficit hyperactivity disorder (ADHD). Noradrenergic systems are critical to higher brain functions such as attention and executive function, which are defective in ADHD. The clinical efficacy of medications that target NET also supports its role in the etiology of ADHD. Here, we have applied a dense mapping strategy to capture all genetic variations within the NET gene in a large number of ADHD families (474 trios). As a result, we found association of the same alleles from two single-nucleotide polymorphisms (rs3785143 and rs11568324) previously identified in another large-scale ADHD genetic study (International Multisite ADHD Geneproject). Furthermore, the effect sizes were consistent across both studies. This is the first time that identical alleles of NET from different studies were implicated, and thus our report provides further evidence that the NET gene is involved in the etiology of ADHD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Faraone S, Sergeant J, Gillberg C, Biederman J . The worldwide prevalence of ADHD: is it an American condition? World Psychiatry 2003; 2: 104–113.
Gaub M, Carlson CL . Gender differences in ADHD: a meta-analysis and critical review. J Am Acad Child Adolesc Psychiatry 1997; 36: 1036–1045.
Biederman J, Faraone SV . Attention-deficit hyperactivity disorder. Lancet 2005; 366: 237–248.
Cantwell DP . Attention deficit disorder: a review of the past 10 years. J Am Acad Child Adolesc Psychiatry 1996; 35: 978–987.
Faraone SV, Doyle AE . The nature and heritability of attention-deficit/hyperactivity disorder. Child Adolesc Psychiatr Clin N Am 2001; 10: 299–316, viii–ix.
Faraone SV . Genetics of adult attention-deficit/hyperactivity disorder. Psychiatr Clin N Am 2004; 27: 303–321.
Biederman J, Faraone SV, Keenan K, Knee D, Tsuang MT . Family-genetic and psychosocial risk factors in DSM-III attention deficit disorder. J Am Acad Child Adolesc Psychiatry 1990; 29: 526–533.
Biederman J, Faraone SV, Keenan K, Benjamin J, Krifcher B, Moore C et al. Further evidence for family-genetic risk factors in attention deficit hyperactivity disorder. Patterns of comorbidity in probands and relatives psychiatrically and pediatrically referred samples. Arch Gen Psychiatry 1992; 49: 728–738.
Thapar A, Hervas A, McGuffin P . Childhood hyperactivity scores are highly heritable and show sibling competition effects: twin study evidence. Behav Genet 1995; 25: 537–544.
Levy F, Hay DA, McStephen M, Wood C, Waldman I . Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. J Am Acad Child Adolesc Psychiatry 1997; 36: 737–744.
Smalley SL . Genetic influences in childhood-onset psychiatric disorders: autism and attention-deficit/hyperactivity disorder. Am J Hum Genet 1997; 60: 1276–1282.
Thapar A, Harrington R, Ross K, McGuffin P . Does the definition of ADHD affect heritability? J Am Acad Child Adolesc Psychiatry 2000; 39: 1528–1536.
Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA et al. Molecular genetics of attention deficit hyperactivity disorder. Biol Psychiatry 2005; 57: 1313–1323.
Berridge CW, Waterhouse BD . The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain Res Brain Res Rev 2003; 42: 33–84.
Arnsten AF, Li BM . Neurobiology of executive functions: catecholamine influences on prefrontal cortical functions. Biol Psychiatry 2005; 57: 1377–1384.
Moron JA, Brockington A, Wise RA, Rocha BA, Hope BT . Dopamine uptake through the norepinephrine transporter in brain regions with low levels of the dopamine transporter: evidence from knock-out mouse lines. J Neurosci 2002; 22: 389–395.
Biederman J, Spencer TJ . Genetics of childhood disorders: XIX. ADHD, Part 3: is ADHD a noradrenergic disorder? J Am Acad Child Adolesc Psychiatry 2000; 39: 1330–1333.
Barr CL, Kroft J, Feng Y, Wigg K, Roberts W, Malone M et al. The norepinephrine transporter gene and attention-deficit hyperactivity disorder. Am J Med Genet 2002; 114: 255–259.
De Luca V, Muglia P, Jain U, Kennedy JL . No evidence of linkage or association between the norepinephrine transporter (NET) gene MnlI polymorphism and adult ADHD. Am J Med Genet B Neuropsychiatr Genet 2004; 124: 38–40.
McEvoy B, Hawi Z, Fitzgerald M, Gill M . No evidence of linkage or association between the norepinephrine transporter (NET) gene polymorphisms and ADHD in the Irish population. Am J Med Genet 2002; 114: 665–666.
Xu X, Knight J, Brookes K, Mill J, Sham P, Craig I et al. DNA pooling analysis of 21 norepinephrine transporter gene SNPs with attention deficit hyperactivity disorder: no evidence for association. Am J Med Genet B Neuropsychiatr Genet 2005; 134: 115–118.
Bobb AJ, Addington AM, Sidransky E, Gornick MC, Lerch JP, Greenstein DK et al. Support for association between ADHD and two candidate genes: NET1 and DRD1. Am J Med Genet B Neuropsychiatr Genet 2005; 134: 67–72.
Kim CH, Hahn MK, Joung Y, Anderson SL, Steele AH, Mazei-Robinson MS et al. A polymorphism in the norepinephrine transporter gene alters promoter activity and is associated with attention-deficit hyperactivity disorder. Proc Natl Acad Sci USA 2006; 103: 19164–19169.
Yang L, Wang YF, Li J, Faraone SV . Association of norepinephrine transporter gene with methylphenidate response. J Am Acad Child Adolesc Psychiatry 2004; 43: 1154–1158.
Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry 2006; 11: 934–953.
Biederman J, Faraone SV, Mick E, Williamson S, Wilens TE, Spencer TJ et al. Clinical correlates of ADHD in females: findings from a large group of girls ascertained from pediatric and psychiatric referral sources. J Am Acad Child Adolesc Psychiatry 1999; 38: 966–975.
Orvaschel H, Puig-Antich J . Schedule for Affective Disorders and Schizophrenia for School-Age Children: Epidemiologic Version. Nova University: Fort Lauderdale, FL, 1987.
Spitzer RL, Williams JB, Gibbon M, First MB . Structured Clinical Interview for DSM-III-R: Non-Patient Edition (SCID-NP, Version 1.0). American Psychiatric Press: Washington, DC, 1990.
Faraone SV, Biederman J, Milberger S . How reliable are maternal reports of their children's psychopathology? One-year recall of psychiatric diagnoses of ADHD children. J Am Acad Child Adolesc Psychiatry 1995; 34: 1001–1008.
de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D . Efficiency and power in genetic association studies. Nat Genet 2005; 37: 1217–1223.
Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G et al. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Brain-derived neutrophic factor. Mol Psychiatry 2002; 7: 579–593.
Dudbridge F . Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 2003; 25: 115–121.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 2007; 81: 559–575.
Lowe N, Kirley A, Hawi Z, Sham P, Wickham H, Kratochvil CJ et al. Joint analysis of the DRD5 marker concludes association with attention-deficit/hyperactivity disorder confined to the predominantly inattentive and combined subtypes. Am J Hum Genet 2004; 74: 348–356.
Maher BS, Marazita ML, Ferrell RE, Vanyukov MM . Dopamine system genes and attention deficit hyperactivity disorder: a meta-analysis. Psychiatr Genet 2002; 12: 207–215.
Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry 2005; 57: 1313–1323.
Kim H, Lim SW, Kim S, Kim JW, Chang YH, Carroll BJ et al. Monoamine transporter gene polymorphisms and antidepressant response in koreans with late-life depression. JAMA 2006; 296: 1609–1618.
Lin PI, Vance JM, Pericak-Vance MA, Martin ER . No gene is an island: the flip-flop phenomenon. Am J Hum Genet 2007; 80: 531–538.
Kim CH, Kim HS, Cubells JF, Kim KS . A previously undescribed intron and extensive 5′ upstream sequence, but not Phox2a-mediated transactivation, are necessary for high level cell type-specific expression of the human norepinephrine transporter gene. J Biol Chem 1999; 274: 6507–6518.
Hahn MK, Mazei-Robison MS, Blakely RD . Single nucleotide polymorphisms in the human norepinephrine transporter gene affect expression, trafficking, antidepressant interaction, and protein kinase C regulation. Mol Pharmacol 2005; 68: 457–466.
Acknowledgements
We are grateful for the families and individuals who participated in this study. We thank Dr Keeley Brookes and Professor Philip Asherson for sharing their data with us, and Brian Galloway for technical assistance. This work was supported by National Institutes of Health (NIH) Grants HD37694, HD37999 and MH66877 to SVF and NARSAD Young Investigator Award to JWK. JWK is an NARSAD Sidney R Baer Jr Foundation Investigator.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, J., Biederman, J., McGrath, C. et al. Further evidence of association between two NET single-nucleotide polymorphisms with ADHD. Mol Psychiatry 13, 624–630 (2008). https://doi.org/10.1038/sj.mp.4002090
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.mp.4002090
Keywords
This article is cited by
-
Adrenergic neurotransmitter system transporter and receptor genes associated with atomoxetine response in attention-deficit hyperactivity disorder children
Journal of Neural Transmission (2013)
-
Possible effect of norepinephrine transporter polymorphisms on methylphenidate-induced changes in neuropsychological function in attention-deficit hyperactivity disorder
Behavioral and Brain Functions (2012)
-
The 1287 G/A polymorphism of the Norepinephrine Transporter gene (NET) is involved in Commission Errors in Korean children with Attention Deficit Hyperactivity Disorder
Behavioral and Brain Functions (2011)
-
No evidence for association between a functional promoter variant of the Norepinephrine Transporter gene SLC6A2 and ADHD in a family-based sample
ADHD Attention Deficit and Hyperactivity Disorders (2011)
-
Molecular genetics of attention-deficit/hyperactivity disorder: an overview
European Child & Adolescent Psychiatry (2010)