Abstract
Alterations in the mismatch repair genes (hMLH1 and hMSH2) play an important role in the development of microsatellite instability in sporadic endometrial cancer. Tissue microarray technology allows molecular profiling of tumor samples at the DNA, RNA, and protein levels. We analyzed hMLH1 and hMSH2 expression by immunohistochemistry in a group of atypical endometrial hyperplasias (n = 10), endometrioid endometrial carcinomas (n = 58), and nonendometrioid endometrial carcinomas (n = 27) on tissue microarray. The results were correlated with microsatellite instability status as evaluated by BAT-25 and BAT-26. Overall, 29.4% of lesions showed microsatellite instability. Loss of nuclear hMLH1 and hMSH2 protein expression was seen in 22.3% and 6.5% of cases, respectively. Immunohistochemistry for hMLH1 and hMSH2 showed lack of protein expression in 64% and 16.6% of microsatellite instability–positive endometrial lesions, respectively. Taken together, hMLH1 or hMSH2 protein expression was absent in 18 of 24 microsatellite instability–positive cases (75% sensitivity). A high level of concordance was found between immunohistochemistry for hMLH1 and hMSH2 and microsatellite instability status evaluated by BAT-25 and BAT-26 (κ value of 0.7). Of the 57 cases found to be microsatellite instability negative, 53 showed normal expression of both proteins (93% specificity). The observed predictive value of absence of expression of hMLH1 for predicting microsatellite instability–positive status was 82%. The predictive value of normal expression of both proteins for predicting microsatellite instability–negative status was 90%These results are consistent with those previously reported in whole tissue sections. Therefore, immunohistochemical analysis of hMLH1 and hMSH2 expression on tissue microarray provides an accurate technique for screening for tumors with microsatellite instability. Tissue microarrays represent an ideal approach for comparing different diagnostic or predictive markers with one another in consecutive tissue microarray sections.
Similar content being viewed by others
Main
Endometrial cancer is the most commonly diagnosed cancer of the female reproductive tract in the United States and other Western countries (1). Although several genes may be altered in these cancers (2), the molecular events in the development of endometrial carcinoma remain poorly defined. Microsatellites are short tandem repeat sequences of 1–6 base pairs, occurring throughout the genome. Because of their repetitive nature, they are prone to errors caused during replication of the DNA. Therefore, alterations in the length of microsatellite alleles in tumor DNA, compared with constitutive DNA from the same individual, represent a form of replication error referred to as microsatellite instability (3). Microsatellite instability is known to accompany defects in at least six genes: human mutL homologue 1 (hMLH1), human mutS homologue 2 (hMSH2), hMSH6, hMSH3, hPMS1, and PMS2. The instauration of microsatellite instability, the so-called mutator phenotype, in one cell has important molecular implications. The microsatellite instability–associated mismatch repair deficiency leads to accumulation of myriad mutations in coding and noncoding DNA sequences. Some small short-tandem repeats, like mononucleotide repeats, are sometimes located within the coding sequence of some important genes (such as transforming growth factor β receptor type II, BAX, insulin-like growth factor II receptor, hMSH3, and hMSH6), and they may be potential targets in the tumor progression of microsatellite instability–positive tumors (2, 4). Sporadic endometrial carcinomas frequently exhibit microsatellite instability (5, 6), particularly of the endometrioid subtype (7, 8, 9). In sporadic endometrial carcinoma, the presence of microsatellite instability is associated with a lack of hMLH1 function, frequently due to hypermethylation of the hMLH1 promoter (10).
The current gold standard for assessing tumor DNA mismatch repair competency is molecular microsatellite instability testing. This is a labor-intensive test that involves extracting and analyzing DNA from both tumor and normal tissue excised at surgery. The DNA is subjected to polymerase chain reaction amplification of several different chromosomal loci that compare microsatellites, running the polymerase chain reaction products through a gel to separate DNA fragments by size, comparing the tumor-normal pairs, and scoring the differences between the two.
However, some recent studies have also shown a good correlation between microsatellite instability analysis and mismatch repair gene protein immunoexpression (11, 12). In this sense, Leach and colleagues (13) and Thibodeau and colleagues (14) reported the use of monoclonal antibodies directed against hMLH1 and hMSH2 in the immunohistochemical analysis of colorectal carcinomas. Subsequent reports described immunohistochemistry of hMLH1 and hMSH2 in sporadic and hereditary nonpolyposis colorectal carcinomas with varying results. In endometrial carcinomas, a decrease in the amount of immunohistochemical staining for hMLH1 and hMSH2 has also been shown to be associated with the presence of microsatellite instability (11, 15, 16). These results have suggested to some investigators that the immunodetection of the gene products might be an easier and rapid method for the identification of tumors of the mutator phenotype and can be used as a prescreening method for the actual mutation status of the mismatch repair system genes (12). Other investigators concluded, however, that immunohistochemistry cannot replace microsatellite instability analysis as a prescreening method because of lower sensitivity (17). Until now, experience seemed to suggest that hMSH2 antibody staining is technically reliable, shows nuclear staining that is easy to interpret, and correlates well with microsatellite instability. In contrast, the hMLH1 antibodies yield a more variable, sometimes patchy staining, often with strong background that is difficult to interpret, and therefore their staining tends not to correlate well with microsatellite instability status (18). Moreover, many of the studies on the value of immunohistochemistry published so far were hampered by a small number of tumors associated with a known mismatch repair system gene mutation.
Immunohistochemistry for diagnostic purposes is usually performed on whole slides. However, this conventional approach of subjecting hundreds of separate tissue sections to immunohistochemical staining is time-consuming and expensive. Thus, there is now substantial interest in developing high-throughput molecular pathology techniques. Tissue microarrays recently have been described by Kononen et al. (19) as being a novel modification of the original method of a multitumor block proposed by Battifora in 1986 (20). The tissue microarray technique allows immunohistochemical analysis of hundreds of samples simultaneously (19, 21, 22). This technology greatly increases the efficiency of tissue-based research. The technique has been demonstrated to be effective and applicable to various tumor types, but methodological evaluations are scarce (23, 24), and therefore, validation studies are necessary to compare tissue microarray cores and standard whole sections.
Previous immunohistochemical studies of endometrial cancer have used whole tissue sections, but, to our knowledge, the use of tissue microarrays has not been evaluated in endometrial tumors. Our aims were to determine the sensitivity of immunohistochemistry for hMLH1 and hMSH2 on tissue microsarray versus microsatellite testing for determining the competence of the mismatch repair mechanism in a large series of premalignant (atypical hyperplasias) and malignant endometrial lesions and to validate the use of tissue microarray for immunohistochemistry of endometrial lesions and compare it with immunohistochemistry on whole tissue sections.
MATERIALS AND METHODS
Patient Selection
The current study comprised 95 endometrial nonconsecutive lesions obtained from surgical patients from the Hospital Universitario La Paz in Madrid between April 1994 and June 2000: 10 atypical endometrial hyperplasias, 58 endometrioid endometrial carcinomas, and 27 nonendometrioid endometrial carcinomas. None of the lesions classified as atypical endometrial hyperplasias were associated with carcinoma. The clinical and pathological findings of these cases were available. All samples used for this study were fixed in 10% formalin and embedded in paraffin. All immunohistochemical and molecular analyses were carried out in paraffin-embedded samples. Table 1 shows the main clinicopathological features of this series.
Tissue Microarray Construction
Representative areas of the different lesions were carefully selected from hematoxylin- and eosin-stained sections and marked on individual paraffin blocks. Samples were chosen from those cases in which more than one large block of the lesion was available so that the availability of tissue for correlative studies would not be compromised. Discrete regions of the lesions that were not necrotic or fibrotic were marked with black ink to guide the technician in the construction of the tissue array. Two tissue cores (1 mm in diameter) were obtained from each specimen. The tissue cores were precisely arrayed into a new paraffin block using a tissue microarray workstation (Beecher Instruments, Silver Spring, MD), as described elsewhere (19). Briefly, the instrument consists of thin-walled stainless steel needles with an inner diameter of approximately 600 μm and a stylet used to transfer and empty the needle contents. The assembly is held in an X-Y position guide that is manually adjusted by digital micrometers. The final tissue microarray consisted of 190 1-mm-diameter tissue cores, with a spacing of 0.8 mm between core centers. A hematoxylin- and eosin-stained section was reviewed to confirm the presence of morphologically representative areas of the original lesions. A tissue core was informative if ≥50% of the disk contained the lesion (carcinoma or atypical hyperplasia).
Immunohistochemistry
To avoid loss of immunoreactivity, microarray slides and whole tissue sections were processed within 1 week of cutting. Conventional immunohistochemistry on whole tissue sections was performed on 4-μm sections of formalin-fixed, paraffin-embedded tissues. Tissue microarray slides were stained with antibodies against hMLH1 and hMSH2, whereas whole tissue sections were stained only with the antibody against hMLH1. Briefly, whole tissue sections and tissue microarray slides were mounted on charged poly-l-lysine-coated slides. The sections were deparaffinized in xylene and rehydrated through a graded alcohol series to distilled water. The slides were subjected to antigen retrieval by microwave irradiation in 10 mm citrate buffer, pH 6.0, in a 750-W oven for 30 min, at a estimated temperature of 95-97° C. The slides were then cooled to room temperature and washed in phosphate-buffered saline. Endogenous peroxidase activity was blocked by the incubation of the slides in hydrogen peroxide and methanol. For hMLH1, we used clone G168–728 (PharMingen, Hamburg, Germany), raised against full-length human MLH1 protein. For hMSH2 we used clone FE11 (Oncogene Research Products, Cambridge, MA), raised against the COOH-terminal fragment of human MSH2 protein. Both monoclonal antibodies were applied for 24 min at 37° C at a 1% concentration in an automated immunostainer (Dako TechMate 500 Plus; DAKO, Glostrup, Denmark). The antibodies were detected by standard indirect immunoperoxidase procedures (LSAB, DAKO). Diaminobenzidine was used as a chromogen, and light hematoxylin was used as a counterstain.
Analysis of Immunohistochemical Stains
Lesions were evaluated for the presence of nuclear staining. Internal positive control for staining consisted of normal endometrium, interspersed inflammatory cells, and/or endothelium. In negative controls, the primary antibodies were omitted.
Individual cores were scored as positive (showing nuclear staining in at least some cells) or negative. Lesions that demonstrated any evidence of hMLH1 or hMSH2 expression, even in small foci, were considered to be positive for hMLH1 or hMSH2 expression. In whole tissue sections, cases displaying loss of staining in the neoplastic cells in selected, circumscribed areas (clonal loss) with concurrent, unequivocal staining of nuclei of nonneoplastic cells were classified as showing abnormal expression of mismatch repair proteins. Cytoplasmic staining in the absence of nuclear staining was not considered immunopositive. Expression of hMLH1 and hMSH2 was determined by two of the investigators (D.H. and J.P.).
Microsatellite Instability Analysis
The microsatellite instability status of the endometrial lesions included in this study had been characterized elsewhere (25). Briefly, we analyzed two mononucleotide repeats, BAT-26 (within intron 5 of the hMSH2 gene on chromosome 2p22–21) and BAT-25 (in an intron of the c-Kit oncogen on chromosome 4q12). DNA was extracted from formalin-fixed, paraffin-embedded tumor sections. All sections were checked for the presence of malignant tumor cells. Normal control DNA was obtained from normal myometrium or fallopian tube tissue. Primers, PCR amplification conditions, and PCR product analyses were made as described elsewhere (26). We considered a phenotype to be microsatellite instability positive when the tumor had deletions of >2 bp in BAT-26 and BAT-25, when compared with the corresponding alleles in the normal tissue. Our previous experience in endometrial (27) and ovarian cancer (28) using the complete Bethesda panel (29), and that of other investigators (30, 31), indicates that the reliability of both mononucleotide repeats is so high that microsatellite instability status can be predicted in most cases by evaluating exclusively BAT-25 and BAT-26.
Statistical Analysis
Associations between the presence of microsatellite instability, expression of hMLH1 and hMSH2 proteins, and several clinicopathological features were calculated using χ2 contingency tests with Yates correction, or Fisher's exact test. κ test was used to test the concordance between the immunohistochemical analysis of hMLH1 and hMSH2 and the microsatellite status of the cases. A κ value of >0.5 was considered to denote a strong association between the two methods of immunohistochemistry and microsatellite instability analysis. All calculations were made using the SPSS-10 statistical program (SPSS, Inc., Chicago, IL). P values of <.05 were considered statistically significant.
Sensitivity and specificity for immunohistochemical classification for microsatellite instability status was defined using the microsatellite instability results as the gold standard.
Sensitivity was defined as follows:
Sensitivity = true positive/(true positive + false negative),
where true positive means absence of hMLH1 and hMSH2 expression by immunohistochemistry in lesions with microsatellite instability demonstrated by PCR analysis, and false negative means presence of hMLH1 and hMSH2 nuclear expression by immunohistochemistry in lesions with microsatellite instability demonstrated by PCR analysis.
Specificity was calculated as follows:
Specificity = true negative/(true negative + false positive),
where a true-negative case is defined as intact expression of hMLH1 and hMSH2 by immunohistochemistry in microsatellite instability–negative lesions, and false positive means absence of nuclear hMLH1 and hMSH2 protein expression in microsatellite instability–negative lesions.
RESULTS
Clinicopathologic Features of Atypical Endometrial Hyperplasia and Endometrial Carcinoma
The mean age of onset of all carcinomas was 64.5 ± 11.2 years (range, 30–89 y, with a mean age of 62.8 ± 12.1 y for the endometrioid carcinomas and 66 ± 11.2 y for the nonendometrioid carcinomas), and for atypical endometrial hyperplasias, it was 49.7 ± 9.76 (range, 38–67 y). Endometrial carcinomas (n = 85) were classified into two groups: (a) endometrioid type (58 of 85; 68.2%) and (b) nonendometrioid (27 of 85, 31.8%). The latter group included papillary serous carcinomas (n = 12), clear cell carcinomas (n = 4), and mixed carcinomas (n = 11). This later group comprised 5 serous–clear cell carcinomas, 4 endometrioid-serous carcinomas, one endometrioid-serous–clear cell carcinoma, and one endometrioid–clear cell carcinoma. According to the FIGO criteria (32), 30 of 58 (51.7%) endometrioid endometrial carcinomas were grade 1 tumors, 17 of 58 (29.3%) were grade 2 tumors, and 11 (18.9%) of 58 were grade 3 tumors.
Staging information was available in 77 of 85 carcinomas. The majority of the endometrioid carcinomas (n = 43, 79.6%) were classified as FIGO stage I, five tumors (9.3%) were classified as FIGO stage II, and six tumors (11.1%) were classified as FIGO stage III. In the group of the nonendometrioid carcinomas, 11 tumors (47.5%) were classified as FIGO stage I, 5 tumors (21.7%) as FIGO stage II, and 7 tumors (30.4%) as FIGO stage III. The differences in the FIGO stage between both groups of tumors were statistically significant (P = .020; Table 1).
Microsatellite Instability Analysis
Of the 95 samples, 85 were suitable for microsatellite instability analysis. In 10 cases (10.5%), the microsatellite instability analysis could not be performed because of DNA degradation. Overall, 25 endometrial lesions (29.4%) were microsatellite instability positive. Twenty of the 52 (38.5%) endometrioid endometrial carcinomas, 3 of the 26 nonendometrioid endometrial carcinomas (11.5%), and 2 of the 7 atypical endometrial hyperplasias (28.6%) exhibited an microsatellite instability–positive phenotype (Fig. 1C–F). The difference between the groups was statistically significant (P = .048; Table 2). There was a trend to association between microsatellite instability and tumor grade (P = .065), with microsatellite instability being more frequent in grade II (50%) and grade III (60%) than in grade I (23.1%) tumors.
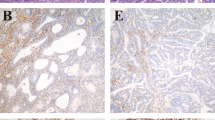
A, tissue core showing absence of hMLH1 expression in a poorly differentiated endometrial carcinoma of the endometrioid type. B, hMLH1 expression is retained in stromal and lymphoid cells. C, in the majority of cases with microsatellite instability, the shortened, unstable alleles could be clearly distinguished from the germ-line allele size. Microsatellite instability was defined by the presence of extra bands that differed by multiple base pairs from their normal counterparts. A new peak (allele) appears in the tumor sample shorter than the germ-line allele size. In the case depicted in (A) and (B), the tumor showed a 6-bp deletion in BAT-25 compared with the normal peak (not shown). D, the same case showed intense nuclear hMSH2 protein expression. E, endometrial carcinoma showing absence of hMSH2 expression. Compare with residual nontumoral endometrial gland. F, this case had a 5-bp deletion in BAT-26 (for explanation, see C).
Immunohistochemical Analysis of hMLH1 and hMSH2
Results for hMLH1 were obtained from 94 samples (188 cores), and those for hMSH2, from 92 samples (184 cores). The loss of information was due to the presence of only normal tissue in two disks and to damaged tissue cores in the remaining six cores. hMLH1 immunohistochemical analysis on whole tissue sections showed loss of nuclear protein expression in 21 of 94 cases (22.3%). No immunohistochemical analysis regarding the expression of hMSH2 on whole slides was performed. Tissue microarray immunohistochemical analysis of the endometrial lesions demonstrated loss of nuclear hMLH1 protein expression in 19.1% of cases (18 of 94; Fig. 1A—B) and loss of hMSH2 in 6.5% (6 of 92). Results of testing of endometrial lesions for microsatellite instability analysis and immunohistochemistry of hMLH1 and hMSH2 are listed in Table 2.
Of the microsatellite instability–positive endometrial lesions, 64% (16 of 25) lacked nuclear hMLH1 protein expression. There was positive staining for hMLH1 in 94.9% (56 of 59) of microsatellite instability–negative endometrial lesions. The difference in hMLH1 protein expression between microsatellite instability–positive and microsatellite instability–negative lesions was statistically significant (P < .0001), with a moderate agreement between both techniques, immunohistochemistry and microsatellite instability analysis (κ value of 0.4; Table 3).
Immunostaining for hMSH2 protein yielded no nuclear staining in 16.6% (4 of 24) microsatellite instability–positive endometrial lesions. There was positive staining for hMSH2 protein in 96.5% (56 of 58) microsatellite instability–negative cases (Fig. 1D). hMSH2 protein expression was significantly different in the two groups of lesions, microsatellite instability positive and microsatellite instability negative (P = .040), with a κ value of 0.08 (Table 3).
Taken together, 18 of the 24 microsatellite instability–positive cases (75%) showed neither hMLH1 nor hMSH2 protein expression by immunohistochemistry, for a sensitivity of 75%. The remaining six microsatellite instability–positive cases showed immunostaining for both hMLH1 and hMSH2 proteins in the tissue core (a false-negative test). These cases corresponded to one atypical complex hyperplasia and five endometrioid carcinomas. Of the 57 cases found to be microsatellite instability negative by microsatellite instability testing, 53 showed normal immunohistochemistry expression of both proteins, for a specificity of 93%. In the remaining four microsatellite instability–negative cases (four endometrioid carcinomas), the tumor cells showed abrogation of both proteins tested in the tissue microarray (a false-positive test). The observed predictive value of absence of expression of either hMLH1 or hMSH2 for predicting a microsatellite instability–positive status was 82%. The predictive value of normal expression of both of these proteins for predicting a microsatellite instability–negative status was 90%. The association of the presence or lack of staining for either protein with the microsatellite instability status in endometrial lesions was statistically significant (P < .0001). In this case, there was a strong agreement between immunohistochemistry and microsatellite instability analysis (κ value of 0.7; Table 3).
When compared, staining on whole tissue sections and tissue microarray was concordant in 91 of 94 (96.8%) cases tested for hMLH1. Three microsatellite instability–positive cases did not show loss of hMLH1 in the tissue microarray core (see above) but showed loss of protein expression in parts of these tumors when studied on whole slides (clonal loss). These cases corresponded to one atypical complex hyperplasia and two endometrioid endometrial carcinomas (one G3, FIGO stage III tumor and one G1, FIGO stage II tumor).
DISCUSSION
Previous studies have shown that microsatellite instability is a common feature in endometrial carcinoma (33). Microsatellite instability was present in 29.4% of the endometrial lesions included in our study. Our results showed a higher frequency of microsatellite instability in endometrial carcinoma of the endometrioid type (38.5%) than has been previously published for this type of tumor. The reasons for this higher percentage may reflect the variability in methods previously used for identification of microsatellite instability. Comparing prior studies reveals a lack of uniformity in defining what constitutes microsatellite instability with regard to the number of markers tested and the specific loci used for testing. We have used two markers (BAT-25 and BAT-26) that are highly reliable and sensitive markers for the detection of tumors with a microsatellite-unstable phenotype (30, 31). In one study of 229 endometrial carcinomas evaluated with a panel of microsatellite markers that included BAT-25 and BAT-26, Basil et al. (34) found an overall incidence of microsatellite instability of 30%, which was higher (34.7%) in the group of endometrioid adenocarcinomas. Similar results were found by Parc et al. (35) in a group of 62 endometrial carcinomas. In a previous analysis of 42 sporadic endometrial carcinomas in Spanish women, Catasus and colleagues (9) found an overall incidence of microsatellite instability in 28% of cases, with a higher incidence of microsatellite instability in endometrioid carcinomas (33%) when compared with nonendometrioid carcinomas (11%), which is in agreement with our results.
In our series, microsatellite instability was correlated with the histological type of the lesion, and there was a trend to association between microsatellite instability and tumor grade. However, the aim of this study was to analyze the expression of hMLH1 and hMLH2 in endometrial lesions on tissue microarrays and to correlate the results obtained with microsatellite instability (BAT-25 and BAT-26) to validate immunohistochemistry on tissue microarrays as a screening method for microsatellite instability in a large series of lesions.
Tissue microarray technology has the potential to significantly accelerate the progress of studies seeking associations between molecular changes and clinical end points (19). Another advantage of this approach is that protein expression can be evaluated in a large number of specimens under uniform test conditions. Another important benefit is that tissue is conserved. One concern about tissue microarray has been the small tissue sample size (0.6–1 mm diameter). However, it is important to realize that the tissue microarray approach has been designed to examine tumor populations and not to survey individual tumors. In this sense, most published tissue microarray studies that have analyzed known markers have confirmed the data obtained previously from conventional studies (21, 26, 36, 37). However, some technical steps are crucial for the construction of a tissue microarray carrying paraffin-embedded tissues so that it can be a source for multiple high-quality sections representing as many arrayed specimens as possible. Tissue loss during sectioning and staining is a common problem of the technique (21, 38, 39). In addition, staining artifacts at the tissue borders are a well-known phenomenon in immunohistochemistry. To minimize the effect that they may have on the staining results, it has been recommended to frame arrayed tumor tissues with one row of normal tissues (24). This methodology has the advantages that the tumor tissues are centered on the array and protected by normal tissues and that peripheral staining artifacts involve normal tissues, not tumor specimens. Moreover, normal tissue may serve as an adequate internal positive control of the immunohistochemical technique.
A close association between microsatellite instability status and immunoreactivity has been reported in colorectal cancer, with retained expression for hMLH1 and hMSH2 in microsatellite-stable tumors and frequent loss of expression in microsatellite-unstable tumors, with a correlation between microsatellite instability status and immunohistochemistry ranging from 75 to 100% (12). In a large multicenter study, Lindor et al. (12) studied microsatellite instability and immunohistochemistry in colorectal tumors from 1144 patients. Immunohistochemical detection of hMLH1 and hMSH2 showed 92.3% sensitivity and 100% specificity for microsatellite instability. That is, all immunohistochemically deficient tumors were microsatellite-unstable tumors, whereas 7.7% of microsatellite unstable tumors did not show a deficiency by immunohistochemistry.
In endometrial cancer, the results of correlation between microsatellite instability analysis and immnunohistochemistry are less conclusive. Staebler et al. (15) found an absence of hMLH1 expression in 7 of 13 (54%) microsatellite instability–positive endometrial carcinomas. Berends et al. (40) detected loss of hMLH1 or hMSH2 protein expression in about 50% of the microsatellite instability–positive endometrial tumors. Parc et al. (35) detected loss of nuclear hMLH1 protein expression in 12 of 21 (57%) microsatellite instability–positive endometrial tumors. In two recent studies, Peiró et al. (16) and Stefansson et al. (41) found an absence of hMLH1 protein expression in 54.5% and 40% of microsatellite instability–positive endometrial carcinomas, respectively. Our immunohistochemical results on tissue microarrays, whereby we found an absence of nuclear hMLH1 protein expression in 64% of microsatellite instability–positive endometrial lesions, are in agreement with those previously reported from whole tissue sections.
The overall low incidence of loss of expression of hMSH2 alone in our study (4 of 24 cases, 16.6%) is in agreement with other published values of sporadic endometrial carcinomas. Katabuchi et al. (42) found a loss of hMSH2 protein in 2/11 (18%) sporadic microsatellite instability–positive endometrial carcinomas. More recently, Staebler et al. (15) reported loss of expression of hMSH2 alone in 3 of 13 (23%) of the microsatellite instability–positive endometrial carcinomas. Another recent but smaller study of endometrial carcinomas in young patients found loss of expression for hMSH2 in 4 of 21 (19%) microsatellite instability–positive cases (35). Stefansson et al. (41) demonstrated lack of nuclear hMSH2 protein expression alone in 19% of endometrial sporadic carcinomas. These data are in accordance with the notion that microsatellite instability is the result of hMLH1 promoter hypermethylation in most sporadic endometrial cancers (10).
A subset of cases (6/24) in our series showed microsatellite instability while expressing both proteins studied (false-negative test). Tumor heterogeneity might explain some of the discrepancies observed between hMLH1 or hMSH2 protein expression and microsatellite instability. In hereditary cases, intratumor heterogeneity should not be a problem because of the fact that loss of mismatch repair gene function, and consequently often abrogation of mismatch repair protein expression, is such an early event that is present in all tumor cells (43). However, in sporadic cases, defects of mismatch repair system genes leading to microsatellite instability may be restricted to subclones of tumor cells. This is exemplified by three microsatellite instability–positive cases in our series that did not show loss of hMLH1 or hMSH2 expression in the tissue microarray core but showed loss of protein expression only in parts of the tumors when studied on whole, conventional sections (clonal loss). In these cases, the tumor areas with negative immunostaining occupied approximately 50% of the tumor section and were clearly demarcated from areas with normal protein expression in tumor cells. Nontumoral cells within these areas showed intense nuclear staining and served as internal positive control. Therefore, the sensitivity of the immunohistochemical analysis of hMLH1 on conventional tissue sections for the detection of mismatch repair system is 87.5% (21 of 25 microsatellite instability–positive cases), which is somewhat higher when compared with the sensitivity of the tissue microarray approach (18 of 25 cases, 75%). These discordant cases illustrate one of the main concerns of the tissue microarray technique, namely if 1-mm core sections of tumor specimens on an array are representative of the whole tumor specimen because of tissue heterogeneity. In addition, if there are discrepancies between array-derived and full section–derived data, this may also lead to different results for clinicopathological correlations based on those data. One recent study by Camp and colleagues (23) described the validation of the tissue microarray technology with regard to the representative value of the arrayed biopsies and the number of cores required per specimen. The results of this study show that 2-fold redundancy can lead to >95% concordance between the two methods. In our study, using duplicate cores for each case on the tissue microarray, the concordance of the immunohistochemical results for hMLH1 between both methods was 96.8%. This is in agreement with a recent study by Hendriks et al. (43), which analyzed the sensitivity of immunohistochemistry for hMLH1, hMSH2, and hMSH6 in a subset of hereditary colorectal tumors from known mismatch repair system gene mutation carriers on whole sections and tissue microarray. A high level of concordance between both methods was found for hMLH1 (85%) and hMSH2 (95%); a somewhat lower concordance level was found for hMSH6 (75%), primarily because of positive staining within the tissue microarray and negative staining with the whole slide immunohistochemistry.
The remaining three false-negative cases might harbor alterations in other mismatch repair genes such as hPMS1, hPMS2, hMSH3, and hMSH6 that were not analyzed in this study. As far as we are aware, only hMSH3 and hPMS2 have been examined for mutations in endometrial carcinomas (44, 45, 46). One study found mutations of the hMSH3 gene in 21% of microsatellite instability–positive endometrial carcinomas but concluded that the mutations were probably a consequence rather than a cause of defective mismatch repair system and microsatellite instability (45). On the other hand, no mutations within the coding region of the hPMS2 gene have been found in endometrioid endometrial carcinomas (46). The role of hMSH6 in relation to different cancer types is less clear. Recently, Stefansson et al. (41) found a pathological staining for hMSH6 in 12.3% of sporadic endometrial carcinomas. Another study of endometrial tumors from hereditary nonpolyposis colorectal carcinomas by de Leeuw et al. (47) also reported that hMSH6 mutation carriers had a phenotype featuring lower microsatellite instability, compared with hMLH1 mutation carriers.
We observed four cases in which either hMLH1 or hMSH2 protein was not expressed but in which the lesion did not exhibit the microsatellite instability phenotype (false-positive test). It is possible that those lesions had not yet accumulated sufficient microsatellite alterations to be detectable with only the panel of two markers used in the present study. In fact, aberrant hMLH1 methylation in microsatellite instability–negative cases has also been described (10).
Although most investigators do not emphasize technical difficulties in the interpretation of the immunohistochemical stains for hMLH1 and hMSH2, these surely exist and may have a significant impact on the final results of the study. In this sense, hMSH2 staining is technically reliable and shows nuclear staining that is easy to interpret. In contrast, the hMLH1 antibodies yield a more variable, sometimes patchy staining, often with strong background, that is more difficult to interpret (18). This is illustrated by a multicenter study of 20 selected colorectal carcinomas that were tested by immunohistochemistry for hMLH1 and hMSH2 at 18 institutions (48). The results of this study showed that staining and interpretation of hMSH2 was successful in most laboratories, whereas use of hMLH1 proved more problematic. However, a significant minority of laboratories demonstrated excellent results, including high discriminatory power for both antibodies. Staining for hMLH1 is difficult when tissue is suboptimally fixed. This is not an uncommon occurrence with archival hysterectomy specimens when the resected organ was submitted intact for pathological evaluation. However, the use of aggressive antigen retrieval methods, including combined microwaving with use of pressure cooker and strict adherence to the interpretation protocols, allows the impact of technical and interpretation errors to be minimized (49, 50).
For the purposes of our study, staining of tumor nuclei for hMLH1 and hMSH2 in the tissue microarray was evaluated as absent (no protein) or present (any evidence). Another possibility is to evaluate the staining semiquantitatively according to the number of positive tumor cells and the staining intensity based on the predominant staining intensity in the lesion (15, 16), but this is less reproducible according to most studies (11, 43, 49, 50). In our opinion, hMLH1 and hMSH2 stains should not be interpreted identically in whole sections and arrays. In fact, staining heterogeneity for mismatch repair proteins within the tumor, which may reflect tumor heterogeneity, may be found in whole tissue sections but rarely in the tissue microarray cores, because of the small size of the sample. This fact, the so-called clonal loss should be considered when interpreting the results of the slides. In any case, we consider that preservation of unequivocal nuclear staining in nonneoplastic tissue (lymphocytes, endometrial glands, endometrial stromal cells, and endothelial cells) should be always used as an internal control requirement before evaluation of tumor is undertaken. Technically unsatisfactory slides with too-strong background, to weak staining, or lack of nuclear staining in positive controls should be rejected.
In summary, in our tissue microarray study, the sensitivity of immunohistochemistry for predicting a lesion with microsatellite instability phenotype was 75%, with a specificity of 93%. In other words, 18 of 24 endometrial lesions with microsatellite instability–positive phenotype did not express either hMLH1 or hMSH2. However, 10.2% (6 of 59) of lesions with normally expressed hMLH1 and/or hMSH2 had a microsatellite instability phenotype. Therefore, in this mixed population of endometrial lesions (endometrial atypical hyperplasias, endometrioid endometrial carcinomas, and nonendometrioid endometrial carcinomas), an abnormal immunohistochemistry result has an 82% predictive value for an microsatellite instability–positive phenotype, and a normal immunohistochemistry test result for these two proteins has a 90% predictive value for an microsatellite instability–negative phenotype. Therefore, the present study shows that it is possible to predict microsatellite instability using immunohistochemical analysis of hMLH1 and hMSH2 in endometrial carcinomas and atypical endometrial hyperplasias on tissue microarray as accurately as in whole tissue sections. Moreover, we found a high level of concordance between immunohistochemistry for hMLH1 and hMSH2 on tissue microarray and microsatellite instability status evaluated by BAT-25 and BAT-26. Thus, the tissue microarray technique appears to offer a relatively convenient, specific, rapid and thus cost-effective method for the immunohistochemical analysis of hMLH1 and hMSH2. Therefore, tissue microarray in immunohistochemical studies offers a time-saving and tissue-preserving technique for studies of multiple biological markers in large tumor series.
References
Landis SH, Murray T, Bolden S, Wingo PA . Cancer statistics, 1998. CA Cancer J Clin 1998; 48: 6–29.
Matias-Guiu X, Catasus L, Bussaglia E, Lagarda H, Garcia A, Pons C, et al. Molecular pathology of endometrial carcinoma and hyperplasia. Hum Pathol 2001; 32: 569–577.
Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulus N, Jen J, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 1993; 75: 1227–1236.
Kinzler KW, Vogelstein B . Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–170.
Risinger JI, Berchuck A, Kohler MF, Watson P, Lynch HT, Boyd J . Genetic instability of microsatellites in endometrial carcinoma. Cancer Res 1993; 53: 5100–5103.
Duggan BD, Felix JC, Mudersprach LI, Turgeman D, Zheng J, Shibata DK . Microsatellite instability in sporadic endometrial carcinoma. J Natl Cancer Inst 1994; 86: 1216–1221.
Helland A, Borresen-Dale AL, Peltomäki P, Hektoen M, Kristensen GB, Nesland JM, et al. Microsatellite instability in cervical and endometrial carcinomas. Int J Cancer 1997; 70: 499–501.
Gurin CC, Federici MG, Kang L, Boyd J . Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res 1999; 59: 462–466.
Catasus L, Machin P, Matias-Guiu X, Prat J . Microsatellite instability in endometrial carcinomas: clinicopathologic correlations in a series of 42 cases. Hum Pathol 1998; 29: 1160–1164.
Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG . MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene 1998; 17: 2413–2417.
Marcus VA, Madlensky L, Gryfe R, Kim H, So K, Millar A, et al. Immunohistochemistry for hMLH1 and hMSH2: a practical test for DNA mismatch repair-deficient tumors. Am J Surg Pathol 1999; 23: 1248–1255.
Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002; 20: 1043–1048.
Leach FS, Polyak K, Burrell M, Johnson KA, Hill D, Dunlop MG, et al. Expression of the human mismatch repair gene hMSH2 in normal and neoplastic tissues. Cancer Res 1996; 56: 235–240.
Thibodeau SN, French AJ, Roche PC, Cunningham JM, Tester DJ, Lindor NM, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res 1996; 56: 4836–4840.
Staebler A, Lax SF, Ellenson LH . Altered expression of hMLH1 and hMSH2 protein in endometrial carcinomas with microsatellite instability. Hum Pathol 2000; 31: 354–358.
Peiró G, Diebold J, Lohse P, Ruebsamen H, Lohse P, Baretton GB, et al. Microsatellite instability, loss of heterozygosity, and loss of hMLH1 and hMSH2 protein expression in endometrial carcinoma. Hum Pathol 2002; 33: 347–354.
Stone JG, Robertson D, Houlston RS . Immunohistochemistry for MSH2 and MLH1: a method for identifying mismatch repair deficient colorectal cancer. J Clin Pathol 2001; 54: 484–487.
de la Chapelle A . Microsatellite instability phenotype of tumors: genotyping or immunohistochemistry?. The jury is still out [editorial]. J Clin Oncol 2002; 20: 897–899.
Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998; 7: 844–847.
Battifora H . The multitumor (sausage) tissue block: novel method for immunohistochemical antibody testing. Lab Invest 1986; 55: 244–248.
Schraml P, Kononen J, Bubendorf L, Moch H, Bissig H, Nocito A, et al. Tissue microarrays for gene amplification surveys in many diffrerent tumor types. Clin Cancer Res 1999; 5: 1966–1975.
Kallioniemi OP, Wagner U, Kononen J, Sauter G . Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet 2001; 10: 657–662.
Camp RL, Charette LA, Rimm DL . Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000; 80: 1943–1949.
Hoos A, Cordon-Cardo C . Tissue microarray profiling of cancer specimens and cell lines: opportunities and limitations. Lab Invest 2001; 81: 1331–1338.
Moreno-Bueno G, Hardisson D, Sánchez C, Sarrio D, Cassia R, Garcia-Rostan G, et al. Abnormalities of the APC/β-catenin pathway in endometrial cancer. Oncogene 2002; 21: 7981–7990.
Moreno-Bueno G, Gamallo C, Perez-Gallego L, Contreras F, Palacios J . β-Catenin expression pattern, β-catenin gene mutations, and microsatellite instability in endometrioid ovarian carcinomas and synchronous endometrial carcinomas. Diagn Mol Pathol 2001; 10: 116–122.
Palacios J, Catasus L, Moreno-Bueno G, Matias-Guiu X, Prat J, Gamallo C . β- and γ-catenin expression in endometrial carcinoma. Relationship with clinicopathological features and microsatellite instability. Virchows Arch 2001; 438: 464–469.
Gras E, Catasus L, Argüelles R, Moreno-Bueno G, Palacios J, Gamallo C, et al. Microsatellite instability, MLH-1 promoter hypermethylation, and frameshift mutations at coding mononucleotide repeat microsatellites in ovarian tumors. Cancer 2001; 92: 2829–2836.
Boland CR, Thibodeau SN, Hamilton SR, Sidranski D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998; 58: 5248–5257.
Perucho M . Correspondence re: Boland CR et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the detection of microsatellite instability in colorectal cancer [letter]. Cancer Res 1999; 59: 249–256.
Zhou XP, Hoang JM, Li YJ, Seruca R, Carneiro F, Sobrinho-Simoes M, et al. Determination of the replication error phenotype in human tumors without the requirement for matching normal DNA by analysis of mononucleotide repeat microsatellites. Genes Chromosomes Cancer 1998; 21: 101–107.
The International Federation of Obstetrics, and Gynecology: 1988 revision. Gynecol Oncol 1989; 35: 125–127.
McDonald ND, Salvensen HB, Ryan A, Iversen OE, Akslen LA, Jacobs IJ . Frequency and prognostic impact of microsatellite instability in a large population-based study of endometrial carcinomas. Cancer Res 2000; 60: 1750–1752.
Basil JB, Goodfellow PJ, Rader JS, Mutch D, Herzog TJ . Clinical significance of microsatellite instability in endometrial carcinoma. Cancer 2000; 89: 1758–1764.
Parc YR, Halling KC, Burgart LJ, McDonnell SK, Schaid DJ, Thibodeau SN, et al. Microsatellite instability and hMLH1/hMSH2 expression in young endometrial carcinoma patients: association with family history and histopathology. Int J Cancer 2000; 86: 60–66.
Moch H, Kononen J, Kallioniemi OP, Sauter G . Tissue microarrays: what will they bring to molecular and anatomic pathology? Adv Anat Pathol 2001; 8: 14–20.
Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, et al. Identification of differently expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res 2000; 60: 6617–6622.
Mucci NR, Akdas G, Manely S, Rubin MA . Neuroendocrine markers in metastatic prostate cancer: evaluation of high throughput tissue microarraysto detect heterogeneous protein expression. Hum Pathol 2000; 31: 406–414.
Richter J, Wagner U, Kononen J, Fijan A, Bruderer J, Schmid U, et al. High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am J Pathol 2000; 157: 787–794.
Berends MJ, Hollema H, Wu Y, van Der Sluis T, Mensink RG, ten Hoor KA, et al. MLH1 and MSH2 protein expression as a pre-screening marker in hereditary and non-hereditary endometrial hyperplasia and cancer. Int J Cancer 2001; 92: 398–403.
Stefansson I, Akslen LA, MacDonald N, Ryan A, Das S, Jacobs IJ, et al. Loss of hMSH2 and hMSH6 expression is frequent in sporadic endometrial carcinomas with microsatellite instability: a population-based study. Clin Cancer Res 2002; 8: 138–143.
Katabuchi H, van Rees B, Lambers AR, Ronnett BM, Blazes MS, Leach FS, et al. Mutations in DNA. mismatch repair genes are not responsible for microsatellite instability in most sporadic endometrial carcinomas. Cancer Res 1995; 55: 5556–5560.
Hendriks Y, Franken P, Dierssen JW, de Leeuw W, Wijnen J, Dreef E, et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol 2003; 162: 469–477.
Risinger JI, Umar A, Boyd J, Berchuck A, Kunkel TA, Barrett JC . Mutation of MSH3 in endometrial cancer and evidence for its functional role in heteroduplex repair. Nat Genet 1996; 14: 102–105.
Swisher EM, Mutch DG, Herzog TJ, Rader JS, Kowalski LD, Elbendary A, et al. Analysis of MSH3 in endometrial cancers with defective DNA mismatch repair. J Soc Gynecol Invest 1998; 5: 210–216.
Basil JB, Swisher EM, Herzog TJ, Rader JS, Elbendary A, Mutch DG, et al. Mutational analysis of the PMS2 gene in sporadic endometrial cancers with microsatellite instability. Gynecol Oncol 1999; 74: 395–399.
de Leeuw WJ, Dierssen J, Vasen HF, Wijnen JT, Kenter CG, Meijers-Heijboer H, et al. Prediction of a mismatch repair gene defect by microsatellite instability and immunohistochemical analysis in endometrial tumors from HNPCC patients. J Pathol 2000; 192: 328–335.
Müller W, Burgart LJ, Krause-Paulus R, Thibodeau SN, Almeida M, Brocker Edmonston T, et al. The reliability of immunohistochemistry as a prescreening method for the diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC). Results of an international collaborative study. Familial Cancer 2001; 1: 87–93.
Manavis J, Gilham P, Davies R, Ruszkiewicz A . The immunohistochemical detection of mismatch repair gene proteins (MLH1, MSH2, MSH6, and PMS2): practical aspects in antigen retrieval and biotin blocking protocols. Appl Immunohistochem Mol Morphol 2003; 11: 73–77.
Ruszkiewicz A, Bennett G, Moore J, Manavis J, Rudzki B, Shen L, et al. Correlation of mismatch repair genes immunohistochemistry and microsatellite instability status in HNPCC-associated tumours. Pathology 2002; 34: 541–547.
Acknowledgements
This study is supported by grants PI020342 and PI020355 from the Fondo de Investigación Sanitaria, Spain and SAF2001-0065. G.M.-B. is a recipient of a postdoctoral research grant from the Centro Nacional de Investigaciones Oncológicas, Spain, and D.S. is a recipient of a BEFI grant from the Fondo de Investigación Sanitaria (01/9132), Spain.
Presented in part at the 92nd Annual Meeting of the United States and Canadian Academy of Pathology (USCAP), Washington, D.C., March 22–28, 2003.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hardisson, D., Moreno-Bueno, G., Sánchez, L. et al. Tissue Microarray Immunohistochemical Expression Analysis of Mismatch Repair (hMLH1 and hMSH2 Genes) in Endometrial Carcinoma and Atypical Endometrial Hyperplasia: Relationship with Microsatellite Instability. Mod Pathol 16, 1148–1158 (2003). https://doi.org/10.1097/01.MP.0000095646.70007.6A
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000095646.70007.6A
Keywords
This article is cited by
-
Frequent loss of mutation-specific mismatch repair protein expression in nonneoplastic endometrium of Lynch syndrome patients
Modern Pathology (2020)
-
T cell-inflamed phenotype and increased Foxp3 expression in infiltrating T-cells of mismatch-repair deficient endometrial cancers
Modern Pathology (2019)
-
Modal variety of microsatellite instability in human endometrial carcinomas
Journal of Cancer Research and Clinical Oncology (2016)
-
hMSH2 is the most commonly mutated MMR gene in a cohort of Greek HNPCC patients
British Journal of Cancer (2005)