Abstract
It has been suggested that gains of chromosomes 7 and 17 and loss of Y occur in renal papillary adenoma and that progression to papillary renal cell carcinoma is marked by gains of additional chromosomes, most frequently 12, 16, and 20. Previous studies have included very few lesions of <5 mm in diameter, a requirement of the present definition of papillary adenoma. Ten papillary adenomas (ranging from 1 to 5 mm in diameter) from autopsy material and 10 surgically resected papillary renal cell carcinomas were studied with fluorescence in situ hybridization in paraffin sections using centromeric probes for chromosomes 7, 12, 16, 17, 20, and Y diluted 1:100 with tDenHyb1 buffer. The signals in 50 to 150 nuclei were counted in each tumor. Controls for all the probes were normal renal tissues from the same patients. Three or more signals per nucleus were frequently observed in papillary adenomas: chromosome 7 (range, 10 to 50%; ≥30% in 9 of 10), 17 (range, 10 to 47%; ≥30% in 7), 16 (range, 1 to 63%; ≥10% in 5), 12 (range, 0 to 32%; ≥10% in 4), and 20 (range, 5 to 49%; ≥10% in 5). Loss of the Y chromosome was observed in 80 to 90% of nuclei in 9 adenomas from males. Three or more signals were frequent in papillary renal cell carcinomas: chromosome 7 (range, 32 to 63%; ≥30% in 10 of 10), 17 (range, 28 to 61%; ≥30% in 7), 16 (range, 0 to 45%; ≥10% in 6), 12 (range, 1 to 37, ≥10% in 5), 20 (range, 2 to 44%; ≥10% in 4). No signal for Y was observed in 12 to 88% (≥81% in 6) of nuclei in 7 carcinomas from males. Statistical analysis showed no difference between adenomas and carcinomas. Gains of chromosomes 7, 17, 16, 12, and 20 and loss of the Y chromosome occur early in the evolution of papillary renal cell neoplasia in tumors that are only a few millimeters in diameter. Progressive gains of these chromosomes do not appear to correlate with the transition from adenoma to carcinoma.
Similar content being viewed by others
INTRODUCTION
In 1997, two consensus conferences, in Heidelberg, Germany, and Rochester, Minnesota, recognized the existence of benign papillary neoplasms of the renal tubular epithelium that are incidental findings which are orders of magnitude more common than clinically recognized papillary renal cell carcinomas. At the Rochester conference, it was recognized that there are no reliable histological differences between these small tumors and many clinically detected papillary renal cell carcinomas, so renal papillary adenoma was defined on the basis of size of ≤5 mm, whereas larger tumors were classified as papillary renal cell carcinomas (1). An alternative proposal was that there should be a cytogenetic definition (2). This was based on reports that papillary renal cell carcinomas have gains of chromosomes 7, 12, 16, 17, and 20, whereas small papillary renal tumors have only gains of chromosomes 7 and 17 and loss of the Y chromosome. These observations also led to the idea that acquisition of additional trisomies of different chromosomes transforms papillary adenoma into papillary renal cell carcinoma. (3, 4). Those studies included very few tumors of ≤5 mm in diameter because tumors of this size are difficult to study by classic cytogenetic methods. To test the hypothesis that gains of chromosomes are limited in renal papillary adenomas, we studied the numbers of chromosomes 7, 17, 12, 16, 20, and Y in 10 papillary adenomas and 10 papillary renal cell carcinomas with interphase cytogenetics using fluorescent in situ hybridization with centromeric probes for chromosomes in paraffin sections.
MATERIAL AND METHODS
Tissue Samples
Ten papillary adenomas, ranging from 1 to 5 mm in diameter (mean, 3 mm) from autopsy material and 10 surgically resected papillary renal cell carcinomas, with adjacent normal renal parenchyma, were selected from the archives of the Department of Pathology and Laboratory Medicine, Indiana University, Indianapolis, Indiana. We selected the papillary renal cell carcinomas to avoid tumors rich in inflammatory cells.
Fluorescence In Situ Hybridization
Sections 5 μm thick were cut from paraffin blocks. The paraffin was removed from the sections with two 10-minute washes in xylene. After hydrating in 100%, 85%, and 70% ethanol solutions (10 min each) and rinsing in distilled water (10 min) and twice in phosphate-buffered saline (pH 7, 10 min each), the slides were fixed in methanol-acetic acid 3:1 for 10 minutes and air dried. The sections were treated in a 2× standard saline citrate solution (SSC) for 15 minutes at 37° C and then dehydrated in consecutive 70%, 85%, and 100% ethanol solutions for 1 minute each and then dried. The sections were bathed in 0.1 mm citric acid (pH 6) solution at 85° C for 1 hour. Then they were again dehydrated in a series of ethanol solutions and dried. The tissue was digested by applying 0.75 mL of pepsin (Sigma, St. Louis, MO) solution (4 mg/mL in 0.9% NaCl, pH 1.5) to each slide and incubating the slides in a humidified box for 30 minutes at 37° C. The slides were rinsed with distilled water for a few seconds, dehydrated again in graded ethanol solutions, and dried. Centromeric probes for chromosomes 7, 17, 12, 16, 20, and Y (Vysis) were used. Each probe was diluted 1:100 in tDenHyb1 buffer (Insitus, Albuquerque, NM). Ten microliters of diluted probe was applied to each slide, and coverslips were placed over the slides. Denaturation was achieved by incubating the slides at 80° C for 10 minutes in a humidified box, then hybridization was done at 37° C for 3 hours. The coverslips were then removed, and the slides were immersed at room temperature in 0.5× SSC for 2 minutes, in 50% formamide/1× SSC for 5 minutes, and in 2× SSC for 2 minutes. The slides were air-dried and counterstained with 10 μL DAPI/Antifade (DAPI in Fluorguard, 0.5 μg/mL; Insitus).
The slides were examined using an Olympus IX-50 microscope (Olympus, Tokyo, Japan) with filters: blue filter cube (Arcturus, Mountain View, CA) for SpectrumGreen (centromeric probes for chromosomes 7, 12, 17, and Y) Vysis probes, SpectrumOrange Filter (Chroma, Brattleboro, VT) for SpectrumOrange (centromeric probes for chromosomes 16 and 20) Vysis probes, and the UV Filter (Chroma) for the DAPI nuclear counterstain. The signals were recorded with a CCD camera.
In Situ Hybridization Analysis
The criteria for evaluating the fluorescence in situ hybridization signals were adapted from Hopman et al. (5, 6) From 100 to 200 nuclei were scored for alpha-satellite signals observed with the fluorescence microscope at 400× magnification. As much as possible, signals from solitary nuclei were counted, but groups of two or three adjacent but not overlapping nuclei were occasionally included in the counts. Nuclei were counted when the entire nuclear circumference had a round-to-oval contour and showed no evidence of fragmentation. Two signals of the same size in close proximity, not connected by a link, were counted as two signals. A diffuse signal was regarded as a signal if it was contiguous and within an acceptable boundary. Two small signals connected by a visible link were counted as one signal. Overlapping nuclei and nuclei with uncertain signals were not counted. There was no significant variation in hybridization efficiency when different areas of the slides were examined. The number of signals visualized in nuclei were tabulated from areas of the slides in which nuclear overlap was minimal.
Statistical Analysis
SAS software, version 8 (Cary, NC) was used for analysis. Fisher’s exact test was used to determine whether a significant association exists between each tumor type (papillary adenoma, papillary renal cell carcinoma) and three or more signals for each of the five chromosomes. A Cochran-Mantel-Haenszel test was also done to test for an overall association between tumor type and three or more signals, stratified by chromosome.
RESULTS
Clinicopathologic Features
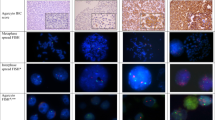
The male-female ratio was 7:1 for renal papillary adenomas and 7:3 for papillary renal cell carcinomas (Table 1). All papillary renal adenomas were ≤5 mm in maximum diameter, ranging from 1 to 5 mm (mean = 3 mm). Most of the carcinomas were >4 cm in diameter, and several were Nuclear Grade 3 or 4. Adenomas and carcinomas of type 1 and type 2 were included in the study (Fig. 1). Five of the adenomas were from two patients who had multiple papillary adenomas.
Histologic features of papillary adenomas and papillary renal cell carcinomas. A, papillary adenoma type 1, a 1-mm tumor within the renal cortex. B, papillary adenoma type 2, a 4-mm subcapsular tumor. C, papillary renal cell carcinoma, type 1, nuclear grade 3. D, papillary renal cell carcinoma, type 2, nuclear grade 4.
Fluorescence In Situ Hybridization
In normal epithelial cells in the renal tubules, nuclei with three or four signals were occasionally seen but in no sample exceeded 12% of the total (Table 2), whereas signal loss always remained at <20% of the total nuclei counted (Fig. 2A).
Three or more signals (Fig. 2B) were frequently observed in papillary adenomas (Table 3): chromosomes 7 (range, 10 to 50%; ≥30% in 9 of 10), 17 (range, 10 to 47%; ≥30% in 7), 16 (range, 1 to 63%; ≥10% in 5), 12 (range, 0 to 32%; ≥10% in 4), and 20 (range, 5 to 49%; ≥10% in 5). No signal for Y was observed in 80 to 90% of nuclei in 9 adenomas from males. Three or more signals were frequent in papillary carcinomas: chromosomes 7 (range, 32 to 63%; ≥30% in 10 of 10), 17 (range, 28 to 61%; ≥30% in 7), 16 (range, 0 to 45%; ≥10% in 6), 12 (range, 1 to 37, ≥10% in 5), and 20 (range, 2 to 44%; ≥10% in 4). No signal for Y was observed in 12 to 88% (≥81% in 6) of nuclei in seven carcinomas from males.
Statistical Analysis
There was no significant difference between renal papillary adenoma and papillary renal cell carcinoma in association with the presence of three or more signals for any chromosome, nor for three or more signals overall.
DISCUSSION
In this study we found that (1) gains of chromosomes 12, 16, and 20 are present in small papillary adenomas; (2) the frequencies of gains of chromosomes 7, 17, 16, 12, 20, and loss of the Y chromosome are similar in both adenomas and carcinomas; (3) no difference in the frequencies of gains of chromosomes 7, 17, 12, 16, and 20 and loss of chromosome Y was found between type 1 and 2 papillary renal cell carcinomas, recognizing that the number of tumors was small; and (4) the epithelium of normal renal tubules adjacent to solitary or multicentric papillary neoplasms shows neither significant gains of chromosomes 7, 17, 16, 12, and 20 nor loss of the Y chromosome.
In 1997, renal papillary adenoma was defined as a lesion ≤5 mm in diameter, with a tubular or a tubulopapillary architecture and not resembling clear, chromophobe, or collecting duct carcinomas (1). The most important criterion distinguishing it from papillary renal cell carcinoma was size, based on data from autopsy studies (7, 8, 9).
The cytogenetic features of papillary renal cell carcinomas have been studied extensively, including a number of small tumors (10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23), but little is known of the cytogenetics of renal papillary adenomas (3, 4, 15, 17). Kovacs et al.(3) and Dal Cin et al. (4) found that gains of chromosomes 7 and 17 and loss of Y occur in renal papillary adenoma. Gains of additional chromosomes, most frequently 12, 16, and 20, were suggested as responsible for the progression to papillary carcinoma. Henke and Erbersdobler (15) recently found that 4 of 17 (24%) renal papillary adenomas (all ≤5 mm in diameter) had three signals in >10% of nuclei for one or more of chromosomes 12, 16, and 20. These data agree with our results, indicating that gains of chromosomes in addition to those of 7 and 17 are frequent in papillary adenomas. Our data show that gains of chromosomes 7, 17, 16, 12, 20 and loss of the Y chromosome are common to both papillary renal cell carcinoma and papillary adenoma. These chromosomal aberrations occur in small tumors of a few millimeters in diameter, early in the evolution of papillary renal cell neoplasia.
The alternative possibility that these small papillary tumors are small carcinomas must be considered. This seems improbable to us because of the tremendously greater numbers of papillary adenomas than clinically detected papillary adenocarcinomas. The numerical relationship is similar to the relationship between colonic tubular adenomas and adenocarcinomas. Combining the chromosome data with the numerical relationship suggests that the early gains of chromosomes may be necessary for the initiation of papillary neoplasia and that papillary adenomas are precursors to papillary renal cell carcinomas, requiring additional genetic changes to acquire invasive and metastatic capabilities.
Previous efforts to study chromosome numbers in small, early papillary renal cell neoplasms have been impeded by methodologic difficulties. Because interphase fluorescent in situ hybridization consumes little tissue and is performed in situ, it facilitates the analysis of small lesions of the renal parenchyma. It permits distinction between normal renal tubules and neoplasm and between the neoplastic epithelial cells and the macrophages in the cores of the papillae in some tumors. Because a single 4- to 5-μm paraffin section is all that is required to analyze for a chromosome, it is possible to study the gain or loss of multiple chromosomes in even a 2- or 3-mm tumor, which is difficult or impossible using traditional cytogenetic methods.
Recently, differences in the frequency of gains of chromosomes between type 1 and type 2 papillary renal cell carcinomas (24) have been found by comparative genomic hybridization (10, 14). Gains of chromosomes 7 and 17 were detected in almost all type 1 tumors, whereas they were considerably less frequent in type 2 papillary renal cell carcinoma. In the limited population of our interphase cytogenetic study, type 1 and type 2 papillary renal cell carcinomas (24) do not appear to have different numbers of the chromosomes we studied.
In conclusion, our data indicate that gains of chromosomes 12, 16, and 20 occur early in the evolution of papillary renal cell neoplasia and do not seem to correlate with the transition from adenoma and carcinoma.
References
Störkel S, Eble JN, Adlakha K, et al. Classification of renal cell carcinoma, workgroup 1. Cancer 1997; 80: 987–989.
Kovacs G, Akhtar M, Beckwith JB, et al. The Heidelberg classification of renal cell tumours. J Pathol 1997; 183: 131–133.
Kovacs G, Fuzesi L, Emanuel A, Kung H-F . Cytogenetics of papillary renal cell tumors. Genes Chromosomes Cancer 1991; 3: 249–255.
Dal Cin P, Gaeta J, Huben R, et al. Renal cortical tumors, cytogenetic characterization. Am J Clin Pathol 1989; 92: 408–414.
Hopman AHN, Ramaekers FCS, Raap AK, et al. In situ hybridization as a tool to study numerical chromosome aberrations in solid bladder tumors. Histochemistry 1988; 89: 307–316.
Hopman AH, Voorter CE, Ramaekers FC . Detection of genomic changes in cancer by in situ hybridization. Mol Biol Rep 1994; 19: 31–44.
Newcomb WD . The search for truth, with special reference to the frequency of gastric ulcer-cancer and the origin of Grawitz tumours of the kidney. Proc R Soc Med 1936; 30: 113–136.
Xipell JM . The incidence of benign renal nodules (a clinicopathologic study). J Urol 1971; 106: 503–506.
Eble JN, Warfel K . Early human renal cortical epithelial neoplasia. Mod Pathol 1991; 4: 45A.
Jiang F, Richter J, Schraml P, et al. Chromosomal imbalances in papillary renal cell carcinoma: genetic differences between histologic subtypes. Am J Pathol 1998; 153: 1467–1473.
Fischer J, Palmedo G, Von Knobloch R, et al. Duplication and overexpression of the mutant allele of the MET proto-oncogene in multiple hereditary papillary renal cell tumours. Oncogene 1998; 17: 733–739.
Lager DJ, Huston BJ, Timmerman TG, Bonsib SM . Papillary renal tumors, morphologic, cytochemical, and genotypic features. Cancer 1995; 76: 669–673.
Corless CL, Aburatani H, Fletcher JA, et al. Papillary renal cell carcinoma, quantitation of chromosomes 7 and 17 by FISH, analysis of chromosome 3p for LOH, and DNA ploidy. Diagn Mol Pathol 1996; 5: 53–64.
Sanders ME, Mick R, Tomaszewski JE, Barr FG . Unique patterns of allelic imbalance distinguish type 1 from type 2 sporadic papillary renal cell carcinoma. Am J Pathol 2002; 161: 997–1005.
Henke RP, Erbersdobler A . Numerical chromosomal aberrations in papillary renal cortical tumors: relationship with histopathologic features. Virchows Arch 2002; 440: 604–609.
El-Naggar AK, Ro JY, Ensign LG . Papillary renal cell carcinoma: clinical implication of DNA content analysis. Hum Pathol 1993; 24: 316–321.
Farnsworth WV, Cohen C, McCue PA, DeRose PB . DNA analysis of small renal cortical neoplasms, comparison with renal carcinomas. J Urol Pathol 1994; 2: 65–79.
Wilhelm M, Veltman JA, Olshen AB, et al. Array-based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res 2002; 62: 957–960.
Bentz M, Bergerheim USR, Li C, et al. Chromosome imbalances in papillary renal cell carcinoma and first cytogenetic data of familial cases analyzed by comparative genomic hybridization. Cytogenet Cell Genet 1996; 75: 17–21.
Kovacs G . Papillary renal cell carcinoma, a morphologic and cytogenetic study of 11 cases. Am J Pathol 1989; 134: 27–34.
Hughson MD, Dickman K, Bigler SA, Meloni AM, Sandberg AA . Clear-cell and papillary carcinoma of the kidney: an analysis of chromosome 3, 7, and 17 abnormalities by microsatellite amplification, cytogenetics, and fluorescence in situ hybridization. Cancer Genet Cytogenet 1998; 106: 93–104.
Kattar MM, Grignon DJ, Wallis T, et al. Clinicopathologic and interphase cytogenetic analysis of papillary (chromophilic) renal cell carcinoma. Mod Pathol 1997; 10: 1143–1150.
Reutzel D, Mende M, Naumann S, et al. Genomic imbalances in 61 renal cancers from the proximal tubulus detected by comparative genomic hybridization. Cytogenet Cell Genet 2001; 93: 221–227.
Delahunt B, Eble JN . Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol 1997; 10: 537–544.
Acknowledgements
The authors thank Joanne Fyffe, M.S., Division of Biostatistics, Indiana University School of Medicine for performing the statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by Fondazione Cassa di Risparmio di Verona (bando 2001); Diagnostica molecolare in oncologia (2003); Murst ex 60% (2003); and Banco di Sardegna.
Presented in part at the 92nd annual meeting of the U.S. and Canadian Academy of Pathology, March 25, 2003.
Rights and permissions
About this article
Cite this article
Brunelli, M., Eble, J., Zhang, S. et al. Gains of Chromosomes 7, 17, 12, 16, and 20 and Loss of Y Occur Early in the Evolution of Papillary Renal Cell Neoplasia: A Fluorescent In Situ Hybridization Study. Mod Pathol 16, 1053–1059 (2003). https://doi.org/10.1097/01.MP.0000090924.90762.94
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000090924.90762.94
Keywords
This article is cited by
-
Identification of prognostic and therapeutic biomarkers in type 2 papillary renal cell carcinoma
World Journal of Surgical Oncology (2023)
-
Effectiveness of intraoperative cell salvage combined with a modified leucocyte depletion filter in metastatic spine tumour surgery
BMC Anesthesiology (2022)
-
Ten-year follow-up of renal adenomatosis with magnetic resonance imaging: a case report
Journal of Medical Case Reports (2022)
-
Papillary renal cell carcinoma: a single institutional study of 199 cases addressing classification, clinicopathologic and molecular features, and treatment outcome
Modern Pathology (2022)
-
MET alterations in biphasic squamoid alveolar papillary renal cell carcinomas and clinicopathological features
Modern Pathology (2021)