Abstract
The p16 (CDKN2a/INK4a) gene is an important tumor-suppressor gene, involved in the p16/cyclin-dependent kinase/retinoblastoma gene pathway of cell cycle control. The p16 protein is considered to be a negative regulator of the pathway. The gene encodes an inhibitor of cyclin-dependent kinases 4 and 6, which regulate the phosphorylation of retinoblastoma gene and the G1 to S phase transition of the cell cycle. In the present study, p16 gene promoter hypermethylation patterns and p16 protein expression were analyzed in 100 consecutive untreated cases of primary head and neck squamous cell carcinoma by methylation-specific PCR and immunohistochemical staining. The p16 promoter hypermethylation and apparent loss of p16 protein expression were detected in 27% and 74% of head and neck squamous cell carcinoma, respectively. By χ2 test, history of alcohol or tobacco use was significantly correlated with the loss of p16 protein expression (P = .005 and.05, respectively). When patient follow-up data were correlated with various clinical and molecular parameters, tumor size and nodal and clinical stage were the strongest prognostic predictors for disease-free survival (tumor recurrence) and for cause-specific and overall survival in patients with head and neck squamous cell carcinoma. Neither p16 promoter hypermethylation nor apparent loss of p16 protein expression appears to be an independent prognostic factor, although loss of p16 protein may be used to predict overall patient survival in early-stage head and neck squamous cell carcinoma.
Similar content being viewed by others
INTRODUCTION
The development of head and neck squamous cell carcinoma is believed to be a multistep process, in which genetic and epigenetic events accumulate as a result of chronic exposure to environmental carcinogens, resulting in loss of cell cycle control, selected cell growth, and finally the development of cancer.
On chromosome 9p21, the INK4a locus encodes both p16 and p14ARF proteins by alternative splicing (1, 2), in which exon 1α is used by p16 and exon 1β is used by p14ARF. Although the role of p14ARF as a true tumor suppressor has yet to be further characterized, p16 fulfills all the criteria for a bona fide tumor-suppressor gene, even judged with the strictest measures (3). The human p16 protein contains 156 amino acids and was first discovered in a yeast two-hybrid system to detect proteins that interact with human cyclin-dependent kinase 4 (4). The tumor suppressor function of p16 is attributed to its ability to inhibit the catalytic activity of the cyclin-dependent kinase 4–6/cyclin D complex that is required for phosphorylation of retinoblastoma protein (5, 6).
The E2F family of transcription factors is essential for cell cycle progression, and their transcriptional activity is inhibited through the specific binding to retinoblastoma protein b (7). This retinoblastoma protein/E2F complex will release E2F transcriptional factors upon phosphorylation of retinoblastoma protein by cyclin dependent kinase 4–6/cyclin D complex and promote cell cycle progression past the G1/S checkpoint (3, 7). By binding to cyclin-dependent kinases 4 and 6 and inhibiting retinoblastoma protein phosphorylation, p16 prevents the formation of cyclin-dependent kinases 4–6/cyclin D stimulatory complex and promotes the formation of retinoblastoma protein/E2F repressive complex, with the end result of blockage of cell cycle progression at the G1/S checkpoint (3).
In head and neck squamous cell carcinoma, a number of studies have identified three major mechanisms by which p16 gene is inactivated, and these are homozygous deletions, inactivating mutation, and aberrant promoter methylation. In the review by Rocco and Sidransky (3), the p16 inactivation was seen in ≤68% of primary human tumors and was caused by homozygous deletions in 27%, by inactivating mutations in 11%, and by promoter hypermethylation in 30% of the cases respectively.
It has been established that loss of p16 function occurs early in cancer progression (8, 9), and some studies even indicate that p16 loss is the earliest detectable genetic abnormality in head and neck squamous cell carcinoma (10, 11).
Consistent with the results of genetic and epigenetic analyses in head and neck squamous cell carcinoma, the p16 gene expression is frequently lost at the protein levels. The frequency of absent p16 protein expression by immunohistochemical staining method in head and neck squamous cell carcinoma ranges from 55 to 90% in three studies involving 332 cases, with an average frequency of 74% (12, 13, 14).
In the present study, we analyze promoter methylation and protein expression patterns of the p16 gene in 100 patients with primary head and neck squamous cell carcinomas that have not been previously treated by radiation and chemotherapy. The epigenetic alteration and protein expression data are then correlated with various clinical parameters in an attempt to determine the prognostic significance of the p16 gene in head and neck squamous cell carcinoma.
MATERIALS AND METHODS
A total of 100 consecutive untreated cases of head and neck squamous cell carcinoma were gathered from the paraffin block archives in the Department of Pathology at John L. McClellan Memorial Veterans' Hospital, spanning a period from 1993 to 1998. Criteria for tissue selection were as follows: (1) primary surgical resection with curative intent or first-time biopsy for adjuvant therapy; (2) an established diagnosis of squamous cell carcinoma by histomorphology of upper aerodigestive tract (lip, oral cavity, pharynx, and larynx); (3) no prior history of head and neck squamous cell carcinoma; and (4) no prior history of chemotherapy or radiation therapy. The histology of each case was reviewed, and representative tissue sections containing invasive squamous cell carcinoma were selected for immunohistochemical staining and extraction of DNA for promoter methylation analysis. Clinical follow-up was available for all patients until May 2002. Pertinent patient information was retrieved from the Computerized Patient Record System of the Department of Veterans Affairs.
Immunohistochemical Staining for the p16 Protein
Immunohistochemical staining was performed as described elsewhere (15), except that the primary antibody was incubated at the dilution of 1:50 for 1.5 hours at room temperature, using a monoclonal antibody against the p16 protein (Ab-4, clone 16P04 or JC2; NeoMarkers, Fremont, CA). The p16 immunohistochemical staining results were interpreted semiquantitatively as follows: negative, <10% positive cells (−); mostly negative, 10 to 20% positive cells (±); weakly positive, 30 to 40% positive cells (+); moderately positive, 50 to 70% positive cells (++); and strongly positive, 80 to 100% positive cells (+++). The above percentages were determined by visual estimation of the positively stained area as compared with all areas containing tumor cells.
Two pathologists (CLZ and CYF) evaluated the slides independently without prior knowledge of the methylation-specific PCR results. Any discrepancy in the immunohistochemical staining interpretation was resolved by reexamination of the cases by both pathologists. One head and neck squamous cell carcinoma case that showed unequivocal positive reaction in both nucleus and cytoplasm was used as the positive control and was included every time when immunohistochemical staining was performed.
Sample Collection and DNA Extraction
DNA samples were collected using the EX-WAX DNA Extraction Kit (Intergen Co., New York, NY) from five deparaffinized, 5-μm-thick tissue sections from each tissue block. Human placental DNA (Sigma) was used as a negative control, and CpGenome universal methylated human DNA (Intergen Co.) served as a positive control, in which all CpG sites within the genomic DNA are chemically methylated.
Bisulfite Modification of DNA for Methylation-Specific PCR
In DNA samples from head and neck squamous cell carcinoma cases, negative and positive controls were subjected to bisulfite modification before MSP using CpGenome DNA modification Kit (Intergen Co.).
PCR Amplification and Primers
Amplification of the promoter region of the p16 gene was carried out in a Touchgene Gradient Thermal Cycler (Techne Inc., Princeton, NJ) in a 50-μL PCR reaction mixture containing 2 μL of bisulfite-treated genomic DNA, dNTPs (each at 200 μm), primers (50 pmol each per reaction), 2.5 mm MgCl2, and 1.25 U Hotstar Taq (Qiagen, Valencia, CA) in 1× PCR buffer. All reagents were supplied with the Qiagen Hotstar Taq Kit (Qiagen). The only exception was the dNTP mix (Roche Molecular Biochemicals, Indianapolis, IN).
Primers for the p16 gene were designed as follows: 5′-TTA TTA GAG GGT GGG GTG GAT TGT-3′ (sense) and 5′-CAA CCC CAA ACC ACA ACC ATA A-3′ (antisense) for the unmethylated reactions; and 5′-TTA TTA GAG GGT GGG GCG GAT CGC-3′ (sense) and 5′-GAC CCC CGA ACC GCG ACCG TAA-3′ (antisense) for the methylated reactions, as described elsewhere (16, 17). Both primers are purchased from Operon Technologies Inc. (Alameda, CA). The PCR conditions were as follows: initial denaturation and hot start at 95° C for 15 minutes, then 40 cycles consisting of 30 seconds at 95° C, 30 seconds at 60° C (unmethylated reactions) or 65° C (methylated reactions), and 1 minute at 72° C, followed by a final 5-minute extension at 72° C. Positive and negative control DNA samples and controls without DNA were used for each set of PCRs.
Statistical Analysis
Statistical analysis was performed on the entire patient population as well as among different subpopulations. Survival was measured in months from the date of diagnosis to the date of death or to the date of last follow-up. Disease-free survival was the period of time from the initial diagnosis of tumor to the first time of tumor recurrence or the development of second primary cancer of the upper aerodigestive tract. Cause-specific survival was based on death as a direct result of tumor progression as compared with death due to other causes. Overall survival was defined as total death of all causes. The association among various factors, such as p16 promoter methylation, p16 protein expression, and clinical and pathologic parameters was analyzed with a cross-table χ2 test. Survival functions and survival possibility of various prognostic factors were calculated with the Kaplan-Meier method, and the log-rank test was used to compare difference of survival curve between groups. The prognostic covariates analyzed included sex, age, tumor site, tumor size, nodal status, clinical stage, and presence or absence of chemo/radiation therapies. All P values were two-sided. SPSS software (Version 11.0, SPSS Inc., Chicago, IL) was used for all statistical computation.
RESULTS
Of 100 patients, 91 underwent curative surgical resection, and 9, chemoradiation therapies as primary treatment for their tumors. The 91 surgically resected tumors all demonstrated clean margins by pathologic examination. All tumor tissues used in the study were derived from primary tumors without prior radiochemotherapy. Nine patients developed second primary tumors of the upper aerodigestive tract (8 lung and 1 esophageal carcinoma), 38 patients developed local recurrences, and 40 had regional lymph node metastasis. Among 100 patients studied, 69 died, 49 from their head and neck cancer and 20 from other causes. The patient population consisted of all males who ranged in age from 45 to 85 years, with a mean age of 65.8 years. The mean follow-up for all patients was 51.2 months. The median survival based on all causes of death was 49.2 months. Of 31 surviving patients, the mean survival was 67.2 months (range, 8.4 to 144 mo). Of the 47 patients with local recurrence or a second primary tumor, 43 (92%) died, and 41 died of their cancer. Nine patients had more than one recurrence, and all died of their disease. Of the 53 patients without local recurrence or a second primary tumor, 26 (49%) died, and 8 died of their disease. The clinical and treatment characteristics of the patient population are summarized in Table 1.
Among 100 cases of head and neck squamous cell carcinoma analyzed for the p16 promoter methylation by methylation-specific PCR, aberrant p16 promoter hypermethylation was seen in 27 (27%) cases, with the remaining 73 cases (73%) showing no evidence of promoter methylation. Positive and negative controls worked appropriately in each round of PCR reaction. Representative p16 methylation-specific PCR was illustrated in Figure 1.
Analysis of p16 promoter methylation by methylation-specific PCR (MSP). Both methylated-specific and unmethylated-specific primer sets are used. PCR reactions 1, 2, 3, 4, and 5 use DNA samples from five primary HNSCCs. Reaction 6 uses water instead as template for blank control. Reactions P and N use universal methylated human DNA in which all CpG cytosines are methylated (Intergen Co.) and human placental DNA (Sigma) as methylation and unmethylation controls. U, unmethylated reaction; M, methylated reaction. As seen in the image, all DNA samples from primary HNSCC contain unmethylated DNA (U). Samples 1 and 2 contain methylated DNA (M), indicative of the presence of p16 promoter methylation. By contrast, DNA Samples 3, 4, and 5 show no evidence of p16 promoter methylation because of absence of methylated reaction (M).
Protein expression of the p16 gene was analyzed by immunohistochemical staining in all 100 cases. Fifty-four, 15, and 5 cases, respectively were determined to be negative (−), mostly negative (±), and weakly positive (+) for the p16 protein, whereas 5 and 21 cases showed moderate (++) to strong (+++) reactivity for the protein (see Materials and Methods for criteria). Thus, it appears that there are 74 cases (74%) that show apparent loss of p16 protein expression (negative to weakly positive by immunohistochemical staining) and 26 cases (26%) with high levels of protein expression (moderately to strongly positive by immunohistochemical staining). Representative p16 immunohistochemical staining was displayed in Figure 2. Because promoter hypermethylation has been postulated as an important mechanism of gene silencing, correlation of p16 promoter hypermethylation with its protein expression was attempted. Among 27 cases that showed p16 promoter methylation, p16 protein expression was absent in 23 (85%) cases but present in 4 (15%) cases. Among 26 cases that displayed high levels of p16 expression by immunohistochemical staining, p16 promoter methylation was absent in 22 (85%) cases but present in 4 (15%) cases. Fifty-one cases displayed apparent loss of p16 protein (negative to weakly positive by immunohistochemical staining), yet absence of p16 promoter methylation. Overall, the p16 promoter hypermethylation was not significantly correlated with p16 protein expression levels (P = .5).
IHC staining for p16 protein in HNSCC. A, from a case strongly positive for the p16 protein; B, from a case that is negative for the protein. Both cytoplasm and nuclei are strongly positive for the p16 protein in a positive case, with the surrounding inflammatory cells and fibroblasts being completely negative (A). By contrast, neoplastic cells are completely negative for p16 protein in a negative case (B).
The association of molecular, clinical, and treatment characteristics with 2-year disease-free survival was analyzed for the entire patient population (100 patients). The 2-year cutoff was used because >90% of head and neck squamous cell carcinoma recurrences occur within 2 years of initial curative treatment. Neither p16 promoter hypermethylation nor apparent loss of p16 protein expression was correlated significantly with 2-year disease-free survival. Tumors arising in the lip were significantly correlated with increased disease-free survival as compared with tumors from other sites (P = .002), probably because of early detection by self-inspection and early treatment. Factors that demonstrated significant association with decreased disease-free survival were increased tumor size (P < .0001), nodal metastasis (P < .0001), more advanced clinical stage (P < .0001), history of chemotherapy (P < .0001), and radiation therapy (p < .0001).
The association of 5-year cause-specific survival (i.e., patients who did not die of their disease) with molecular, clinical, and treatment characteristics was examined on the entire patient population (100 patients). Neither p16 promoter hypermethylation nor apparent loss of p16 protein expression was correlated significantly with 5-year cause-specific survival. Patients whose tumor arose from the lip and who did not drink experienced increased cause-specific survival (P = .002 and 0.04, respectively). Variables demonstrating significant association with decreased cause-specific survival included increased tumor size (P = .001), nodal metastasis (P = .006), more advanced clinical stage (P = .002), history of chemotherapy (P = .03), and radiation therapy (P = .001).
The correlation of overall survival (i.e., patients who did not die) with molecular, clinical, and treatment characteristics of the entire patient population (100 patients) was also performed. Neither p16 promoter hypermethylation nor apparent loss of p16 protein expression was correlated significantly with overall survival. Potential prognostic factors demonstrating significant association with decreased overall survival included increased tumor size (P = .003), nodal metastasis (P = .02), and more advanced clinical stage (P = .005).
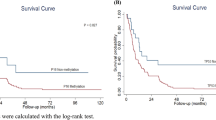
When overall survival was investigated more specifically in different subpopulations by alternatively censoring various tumor, nodal, and clinical stages (Table 2), decreased overall survival was found to be significantly correlated with apparent loss of p16 protein expression in tumors at early stage, as characterized by T1 (P = .008), N0 (P = .05), and clinical Stage I (P = .008) diseases (Table 2).
Neither p16 promoter hypermethylation nor apparent loss of p16 protein expression was correlated significantly with disease-free and cause-specific survival when tumors of early and advanced stages were separately analyzed.
We also determined whether p16 promoter hypermethylation or protein expression levels in the tumors of 100 patients correlated with various clinical, pathologic, and treatment parameters by χ2 test. The presence of p16 promoter hypermethylation or protein expression levels in the primary tumor was not significantly correlated with age ≥65 years, tumor site, nodal status, and chemoradiation therapy. Apparent loss of p16 protein expression was significantly correlated with history of use of tobacco (P = .05) and alcohol (P = .005).
DISCUSSION
In this study, p16 promoter methylation and protein expression analyses were performed in 100 primary head and neck squamous cell carcinoma to determine whether p16 promoter hypermethylation or apparent loss of protein expression could affect other potential prognostic factors, tumor recurrence (disease-free survival), or overall patient survival. Our results indicated that history of tobacco and alcohol use was significantly correlated with apparent loss of p16 protein.
Because tobacco and alcohol are two well-established and significant risk factors in the development of upper aerodigestive tract cancer, and inactivation of p16 tumor suppressor gene is frequently detected in upper aerodigestive tract cancer, several studies have attempted to establish a link between tobacco use and p16 promoter methylation in lung cancer and normal or preinvasive bronchial lesions from smokers (18, 19, 20).
The prevalence of p16 promoter hypermethylation was significantly associated with tobacco exposure in lung (20) and head and neck squamous cell carcinoma (21). In animal studies, smoking increased the frequency of p16 promoter methylation in a dose-dependent pattern (22, 23).
Although our analysis detected significant association of tobacco smoke and alcohol exposure with apparent loss of p16 protein, we failed to establish a similar correlation of p16 promoter hypermethylation with tobacco exposure in head and neck squamous cell carcinoma as demonstrated in lung cancer (20, 23). Such a discrepancy may reflect a different mechanism involved in p16 inactivation between these two types of tobacco-related tumors. In head and neck squamous cell carcinoma, the p16 gene is inactivated by an epigenetic mechanism associated with promoter hypermethylation in <30% of the cases and by genetic factors, such as deletion and mutation in >60% of the cases (3, 24). By contrast, in lung cancer, p16 inactivation is caused by promoter hypermethylation in >70% of the cases and by gene deletion or mutation in <30% of the cases (24).
There appears an excellent agreement between p16 promoter methylation and loss of p16 protein expression in this study. Twenty-three of 27 (85%) cases with p16 promoter hypermethylation showed apparent loss of p16 protein expression, and 22 of 26 (85%) cases with high levels of p16 protein were free of p16 promoter methylation. The lack of overall correlation between p16 promoter methylation and protein expression could be attributed to a large number of cases (51%) in which apparent loss of p16 protein expression was not accompanied by the presence of p16 promoter methylation. Loss of p16 protein expression in these cases may result from aberrant genetic changes, such as homogenous deletion or inactivating mutations, commonly detected in head and neck squamous cell carcinoma (3, 24).
In four cases, tumor cells displayed high levels of p16 protein expression, yet demonstrated p16 promoter hypermethylation (see Results). Such lack of correlation may be due to the heterogeneous p16 immunohistochemical staining patterns in primary head and neck squamous cell carcinoma. In fact, approximately 10% of the tumor cells in all four cases did not express the p16 protein, and this small population of negative cells could very well be the source of methylated DNA detected in these four cases. In future studies, areas with or without p16 protein expression in a given tumor could be separately microdissected using a laser-capture microdissection approach, and DNA samples from these two different areas would then be analyzed for promoter methylation.
Patients who received adjuvant chemoradiation therapy experienced a significantly higher tumor recurrence rate and shorter survival, and this most likely results from the tendency to apply adjuvant therapies to patients with advanced disease. Similar results were obtained in another study involving 91 primary head and neck squamous cell carcinoma (25).
The correlation of p16 inactivation or aberrant genetic or epigenetic changes of the p16 gene with patient survival has been previously investigated in head and neck squamous cell carcinoma (14, 26, 27, 28), with no general consensus. In these studies, p16 alterations have been shown to be associated with decreased patient survival in two studies (14, 28) but demonstrate no difference in patient survival in two other studies (26, 27), underscoring the complex relationship between p16 alterations and prognostic parameters, such as tumor recurrence or patient survival.
In the present study, we investigated how specific molecular markers (p16 promoter methylation or loss of p16 protein expression) affected disease-free, cause-specific, and overall survival in 100 consecutive, untreated cases of head and neck squamous cell carcinoma. With the entire patient population as the basis for statistical analysis, tumor, nodal, and clinical stage remain the strongest prognostic predictors (see Results). Neither p16 promoter methylation nor loss of p16 protein expression can independently predict tumor recurrence and patient survival. However, when a subgroup of patients was analyzed by alternatively censoring tumor, nodal, and clinical stages, apparent loss of p16 protein expression (negative to weakly positive by immunohistochemical staining) was found to be significantly correlated with shortened overall survival in tumors at an early progression stage (T1, N0, and clinical Stage I tumors; Table 2). In this group of patients, the overall probability of surviving 5 years is about 80% in p16-positive tumors and 40% in p16-negative tumors (Table 2). By contrast, in patients with tumors at more advanced stages (T2–4, N1–2, and clinical Stage II-IV tumors), the overall probability of surviving 5 years is similar between p16-positive and p16-negative cases, ranging from 25 to 35% (Table 2).
Because p16 alterations occur early during tumor progression, it is not surprising to find that p16 inactivation primarily influences the patient prognosis in tumors at early progression stage. Similar results were obtained in lung cancer in which p16 promoter methylation was significantly associated only with Stage I tumor but not with tumors of more advanced stages (20). Presumably, with tumors progressing to more advanced stages, additional alterations in other genes critical in tumor progression or invasion accumulate.
In summary, we detected p16 promoter hypermethylation and apparent loss of p16 protein expression in 27% and 74% of head and neck squamous cell carcinoma, respectively. When patient follow-up data were correlated with various clinical and molecular parameters, tumor size, nodal and clinical stage were the strongest prognostic predictors for disease-free survival (tumor recurrence), cause-specific, and overall survival in patients with head and neck squamous cell carcinoma. Even though apparent loss of p16 protein expression may be used to predict overall patient survival at early-stage head and neck squamous cell carcinoma, neither p16 promoter hypermethylation nor apparent loss of p16 protein expression can be used as an independent prognostic factor.
Exposure to tobacco and alcohol enhances the likelihood of p16 inactivation in head and neck squamous cell carcinoma, implying a link between p16 inactivation, the earliest detectable molecular event in the tumor development, and two well-established risk factors in head and neck squamous cell carcinoma.
References
Duro D, Bernard O, Della Valla V, et al. A new type of p16 gene transcript expression in B-cell malignancies. Oncogene 1995; 11: 11–29.
Mao L, Merlo A, Bedi GC, et al. A novel p16 transcript. Cancer Res 1995; 55: 2995–7.
Rocco JW, Sidransky D . p16 (MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res 2001; 264: 42–55.
Ruas M, Peters G . The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta 1998; 1378: F115–77.
Serrano M, Hannon GJ, Beach D . A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993; 366: 704–7.
Serrano M, Lee H, Chin L, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996; 85: 27–37.
Hengstschlager M, Hengstschlager-Ottnad E, Pusch O, et al. The role of p16 in the E2F-dependent thymidine kinase regulation. Oncogene 1996; 12: 1635–43.
Guan RJ, Fu Y, Holt PR, et al. Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology 1999; 116: 1063–71.
Reznikoff CA, Yeager TR, Belair CD, et al. Elevated p16 at senescence and loss of p16 at immortalization in human papillomavirus 16 E6, but not E7, transformed human uroepithelial cells. Cancer Res 1996; 56: 2886–90.
van der Riet P, Nawroz H, Hruban RH, et al. Frequent loss of chromosome 9p21–22 early in head and neck cancer progression. Cancer Res 1994; 54: 1156–8.
Califano J, van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 1996; 56: 2488–92.
Reed AL, Califano J, Cairns P, et al. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res 1996; 56: 3630–3.
Gruttgen A, Reichenzeller M, Junger M, et al. Detailed gene expression analysis but not microsatellite marker analysis of 9p21 reveals differential defects in the INK4a gene locus in the majority of head and neck cancers. J Pathol 2001; 194: 311–7.
Bova RJ, Quinn DI, Nankervis JS, et al. Cyclin D1 and p16INK4a expression predict reduced survival in carcinoma of the anterior tongue. Clin Cancer Res 1999; 5: 2810–9.
Fan C, Liu KL, Huang HY, et al. Frequent allelic imbalance and loss of protein expression of the DNA repair gene hOGG1 in head and neck squamous cell carcinoma. Lab Invest 2001; 81: 1429–38.
Rosas SLB, Koch W, Carvalho MDC, et al. Promoter hypermethylation patterns of p16, 06-methylguanine-DNA methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res 2001; 61: 939–42.
Sanchez-Cespedes M, Esteller M, Wu L, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res 2000; 60: 892–5.
Lamy A, Sesboue R, Bourguignon J, et al. Aberrant methylation of the CDKN2a/p16INK4a gene promoter region in preinvasive bronchial lesions: a prospective study in high-risk patients without invasive cancer. Int J Cancer 2002; 100: 189–93.
Belinsky SA, Palmisano WA, Gilliland FD, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res 2002; 62: 2370–7.
Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 2001; 61: 3419–24.
Hasegawa M, Nelson HH, Peters E, et al. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene 2002; 21: 4231–6.
Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 1998; 95: 11891–6.
Swafford DS, Middleton SK, Palmisano WA, et al. Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol 1997; 17: 1366–74.
Herman JG, Baylin SB . Promoter-region hypermethylation and gene silencing in human cancer. In: Vogt PA, Ja PK, editors. DNA methylation and cancer. 1st ed. New York: Springer-Verlag; 2000. p. 35–50.
Grandis JR, Melhem MF, Gooding WE, et al. Levels of TGFa and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst 1998; 90: 824–32.
Danahey DG, Tobin EJ, Schuller DE, et al. p16 mutation frequency and clinical correlation in head and neck cancer. Acta Otolaryngol (Stockh) 1999; 119: 285–8.
Kwong J, Lo KW, To KF, et al. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res 2002; 8: 131–7.
Bazan V, Zanna I, Migliavacca M, et al. Prognostic significance of p16 alterations and 9p21 loss of heterozygosity in locally advanced laryngeal squamous cell carcinoma. J Cell Physiol 2002; 192: 286–93.
Acknowledgements
This work was supported by research funds from the Departments of Pathology and Otolaryngology, University of Arkansas for Medical Sciences and Biomedical Research Foundation, John L. McClellan Memorial Veteran's Hospital, Little Rock, Arkansas.
Presented at the 92nd Annual Meeting of the United States and Canadian Academy of Pathology, Washington, D.C., March 22–28, 2003.
The authors thank Pamela Ratliff for helping with the p16 immunohistochemical staining.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ai, L., Stephenson, K., Ling, W. et al. The p16 (CDKN2a/INK4a) Tumor-Suppressor Gene in Head and Neck Squamous Cell Carcinoma: A Promoter Methylation and Protein Expression Study in 100 Cases. Mod Pathol 16, 944–950 (2003). https://doi.org/10.1097/01.MP.0000085760.74313.DD
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000085760.74313.DD
Keywords
This article is cited by
-
Preclinical evaluation of the VEGF/Ang2 bispecific nanobody BI 836880 in nasopharyngeal carcinoma models
Investigational New Drugs (2023)
-
A Case of HPV-Associated Oropharyngeal Squamous Cell Carcinoma with Block-Like, Partial Loss of p16 Expression
Head and Neck Pathology (2022)
-
Portrait of DNA methylated genes predictive of poor prognosis in head and neck cancer and the implication for targeted therapy
Scientific Reports (2021)
-
Promoter DNA Methylation and mRNA Expression Level of p16 Gene in Oral Squamous Cell Carcinoma: Correlation with Clinicopathological Characteristics
Pathology & Oncology Research (2019)
-
Disease-relevant signalling-pathways in head and neck cancer: Taspase1’s proteolytic activity fine-tunes TFIIA function
Scientific Reports (2017)