Abstract
To verify the absence of the synovial sarcoma translocation t(X;18) (SYT-SSX) in malignant peripheral nerve sheath tumors, 34 tumor samples from 25 neurofibromatosis type 1 patients were examined in two independent laboratories (Bordeaux, France, and Lausanne, Switzerland) using reverse transcriptase polymerase chain reaction (RT-PCR)-based techniques. RNA was extracted from paraffin blocks using standard methods, reverse transcribed, and conventional (in one laboratory) versus real-time (in the other laboratory) PCR performed. Twenty-seven tumor samples from 19 patients were negative for the t(X;18) in both laboratories; six additional tumors that were t(X;18)-negative in one laboratory gave noninterpretable results in the other, due to lack of internal positive controls; one case was noninterpretable in both places. In conclusion, malignant peripheral nerve sheath tumors in neurofibromatosis type 1 patients do not bear the synovial sarcoma t(X;18) (SYT-SSX). Laboratories that use PCR-based techniques for diagnostic purposes would benefit from quality assurance programs.
Similar content being viewed by others
INTRODUCTION
Most synovial sarcomas bear the translocation t(X;18), which may be detected in formalin-fixed, paraffin-embedded material by reverse transcriptase-polymerase chain reaction (RT-PCR). This technique is particularly useful for consultation cases for which paraffin-embedded material only is available.
Several large series have shown that this method is highly sensitive and specific, with virtually no t(X;18) (SYT-SSX) detected in mesenchymal tumors other than synovial sarcomas (1, 2, 3, 4). This specificity was recently challenged by O'Sullivan et al. (5). These authors detected SYT-SSX fusion gene transcripts not only in synovial sarcomas but also in 15 of 20 malignant peripheral nerve sheath tumors (MPNSTs) as well as in several other types of benign and malignant mesenchymal tumors, thus raising major concerns and controversies regarding t(X;18) specificity. In a letter to the Editor that strongly disagreed with O'Sullivan et al., Ladanyi et al. (6) regretted that the authors did not attempt to confirm their results using alternative techniques such as Southern blotting, conventional cytogenetics, and/or in situ hybridization. In addition, they reported 145 cases of MPNST that always proved to be t(X;18)-negative in several different laboratories using different techniques, a finding that would strongly support their view.
In this article, we report our experience with 34 cases of MPNST that occurred in 25 neurofibromatosis type 1 (NF1) patients. In none of our cases was the t(X;18) (SYT-SSX) detected using two different PCR-based techniques performed in two different laboratories.
MATERIALS AND METHODS
Cases of MPNST occurring exclusively in NF1 patients were retrieved from the files of the Laboratory of Pathology, Institut Bergonié, Bordeaux, France, and the Institute of Pathology, Lausanne, Switzerland. Thirty-four samples from 25 patients were selected. One paraffin block of tumor, at least, was available for molecular testing. Twenty-six samples were fixed in phosphate-buffered formalin and eight in the Holland Bouin fluid (a mixture consisting of water, 9% formaldehyde, 1.35% acetic acid, 3.6g/100 mL picric acid, and 2.25g/100 mL copper acetate [Verbièse, Wasquehal, France]).
In one laboratory (Bordeaux, France), 20 10-μm-thick sections were obtained by cutting paraffin blocks with a microtome. To eliminate any contamination of one case by material from another, the microtome knife was carefully cleaned with absolute alcohol before cutting. In the other laboratory (Lausanne, Switzerland), approximately 50 mg of tumor were removed from paraffin blocks by scraping with a sterile and disposable scalpel. Specific attention was paid to eliminate potential contamination.
RNA extraction protocol was the same in both laboratories (4, 7). Briefly, tissue samples were deparaffinized twice in xylene. After centrifugation, the tissue was washed in ethanol, and the pellet was resuspended with 500 μL of ATL buffer (Qiagen) added with proteinase K and incubated 16 to 48 hours at 60°C. RNA was extracted according to the method of Chomzynski and Sacchi (8) using 1.5 mL of Trizol-LS reagent (Gibco BKL) for 500 μl of cellular lysate. The RNA pellet was resuspended in 50 μl of RNase-free water and stored at −80°C.
Protocol for reverse transcription was the same in Bordeaux and Lausanne (4, 7). Reverse transcription of 5 μg RNA was performed in a total volume of 20 μL with 50 mm Tris-HCl, pH 8,3, 40 mm KCl, 5 mm MgCl2, 0,5% Tween, 0,5 mm dNTP Mix, 10 mm dithiothreitol, specific reverse primer (65 ng SSX-B or 50 ng GAPDH in Bordeaux and 15 ng of β-actin-B in Lausanne), 12 U RNAse inhibitor (Promega), 10U Expand Reverse Transcriptase (Roche Diagnostics). Samples were incubated at 42°C for 1 hour, then at 95°C for 5 minutes.
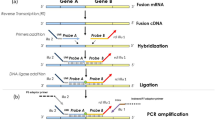
A real time PCR technique was used in one laboratory (Bordeaux, France) (7), and a “conventional” technique was used in the other laboratory (Lausanne, Switzerland) (4). The real-time PCR was carried out using the ABI/PRISM 5700 (Applied Biosystems). PCR reaction was performed in triplicate in a total volume of 50 μL with PCR Core Reagent kit (Applied Biosystems) consisting of 4 mm MgCl2, 0.4 mm of dATP, dCTP, dGTP and 0,8 mm of dUTP, 1,5 U Taq Gold polymerase, 0,5 U Amperase UNG, 4 μL of the cDNA reaction, 65 ng forward primer, 65 ng reverse primer and 5 pmole probe. Thermal cycling conditions were 2 minutes at 50°C for amperase activation, 10 minutes at 95°C for Taq polymerase activation then 50 cycles of 3 PCR steps consisting of 30 seconds at 95°C, 45 seconds at 63°C and 75 seconds at 72°C. GAPDH transcript expression has been studied as a housekeeping gene to control the RNA ability to be reverse transcribed and PCR amplified for all the tumors.
For the conventional PCR method, the amplification reactions (20 μL) were performed, in presence of 2 μL of the cDNA reaction, with an initial incubation step at 95°C for 5 minutes. The amplification profile of the PCR consisted of 35 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 45 seconds, and extension at 72°C for 75 seconds. For each sample, two PCR amplifications were performed in parallel: one contained only the primer set necessary for the amplification of the SYT-SSX fusion gene transcripts, whereas in the second a set of primers at a lower concentration (1 pmole in place of 10 pmoles for the SYT-SSX fusion gene in each 20 μL PCR reaction) for the ubiquitously expressed β-actin gene transcripts were also added. The reaction products were subjected to electrophoresis in 2% agarose gel and visualized by ethidium bromide staining. The RT-PCR procedure was performed at least twice for each sample. For both methods, PCR amplifications were performed with the following primers: STY-A (5′-CAGCAGAGGCCTTATGGATATGA-3′), SSX-B (5′-TTTGTGGGCCAGATG). For PCR amplification of GAPDH, the following primers were used: GAPDH-A (5′ CCA CAT CGC TCA GAC ACC AT 3′), GAPDH-B (5′ CCA GGC GCC CAA TAC G 3′) and for β-actin; β-actin-A (5′-AGGCCAACCGCGAGAAGATGA-3′) and β-actin-B (5′ GCCGTGGTGGTGAAGCTGTAG-3′). For the real-time PCR method, the following probes were used (9): Syno S1 probe (5′ FAM-ATC ATG CCC AAG AAG CCA GCA GAG G 3′) and GAPDH probe (5′ FAM-AAG GTG AAG GTC GGA GTC AAC GGA TTT G 3′).
RESULTS
Clinicopathologic Data
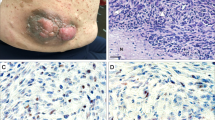
(see Table 1) There were 17 males and eight females, whose ages ranged from 14 to 79 years (mean: 42 years). All patients had clinical features of neurofibromatosis type 1, and for some of them, a familial history was also on records. Tumors were located in lower extremity (n = 4), upper extremity (n = 3), neck (n = 3), trunk wall (n = 6), inner trunk (n = 7), and head (n = 2). From these patients, 34 tumor samples were analyzed, including 20 primary tumors, 12 local recurrences, and two distant metastases.
Histologically, four samples were malignant neurofibromas with an immunopositivity for S100 protein, 22 samples were conventional MPNST with S100 protein reactivity for 17 of them, and eight samples were undifferentiated spindle and/or pleomorphic cell tumors, of which four were S100-positive.
Molecular Findings
Molecular analysis performed in one laboratory (Bordeaux) showed that 30 samples from 21 patients were negative for the t(X;18) (SYT-SSX) in the presence of positive PCR (GAPDH) internal controls (Ct value ranged from 21 to 37, mean: 27). Four tumor samples from four patients (Cases 2, 12, 14, and 17) showed negative results for both SYT-SSX fusion gene transcripts and GAPDH and, thus, were considered noninterpretable. For one of these patients (Case 2) a sample taken from another tumoral event was also negative for the t(X;18) with positive GAPDH PCR internal controls. In the other laboratory (Lausanne, Switzerland), 30 tumor samples from 21 patients were negative for the t(X;18) and positive for β-actin (internal control), whereas four samples from four patients (Cases 14, 18, 22, and 25) were noninterpretable (i.e., lack of amplification of both β-actin and SYT-SSX sequences). Combining the results from the two different laboratories regarding the same tumor population, it appeared that 27 tumor samples from 19 patients were t(X;18)-negative with positive GAPDH/β-actin internal controls, and one sample (Case 14) was noninterpretable due to negative internal controls in both laboratories. Six additional samples from six different patients (Cases 2, 12, 17, 18, 22, and 25) were considered noninterpretable due to negative PCR internal controls obtained in one of the two laboratories (three cases in Bordeaux and three cases in Lausanne).
DISCUSSION
Diagnosing synovial sarcomas may be difficult, and this tumor may be easily confused with a large number of other benign and malignant spindle cell tumors. The demonstration of the translocation t(X;18) (SYT-SSX) in paraffin-embedded material using RT-PCR is now considered a useful and powerful adjunct for the diagnosis of synovial sarcoma due to the high sensitivity and specificity of this method. In a recent study (4) on 250 paraffin-embedded soft-tissue lesions, we obtained PCR products for 221 tumors. t(X;18) (SYT-SSX) fusion gene transcripts were observed in 83 of 86 synovial sarcomas (96% sensitivity) and never detected in any of the 135 tumors of other type that had been examined (100% specificity). Using the same technique on the same kind of material, O'Sullivan et al. (5) found (SYT-SSX) fusion gene transcripts in 29 of 34 (85%) synovial sarcomas, but also in 15 of 20 (75%) MPNSTs, 1 of 4 adult fibrosarcomas, 1 of 10 malignant fibrous histiocytomas, 1 of 7 congenital-infantile fibrosarcomas, and 2 of 3 neurofibromas. Based on these results, the authors concluded that the translocation t(X;18) is not specific for synovial sarcoma and, thus, should be used with great caution in the differential diagnosis of spindle cell tumors.
In a subsequent article on “gold standard” methods for diagnosing soft-tissue tumors (10), the same authors assumed that “molecular genetic testing is an ancillary technique comparable to electron microscopy and immunohistochemistry.” Unfortunately, O'Sullivan et al. (5) did not try to confirm their unexpected and provocative results with techniques other than RT-PCR such as in situ hybridization, which could have been applied on paraffin-embedded material as well. They did not ask another independent laboratory to verify their results on the same samples either. In a letter to the Editor, Ladanyi et al. (6) raised the possibility of contamination during the block cutting procedure or PCR technique based on the fact that none of the 145 cases of MPNST molecularly examined heretofore in different laboratories using different techniques (i.e., RT-PCR, conventional cytogenetics, and FISH) proved to be t(X;18) (SYT-SSX)-positive.
In the current report, we tested 34 samples from 25 cases of MPNST in NF1 patients in two independent laboratories using conventional and real-time PCRs. None of MPNSTs were t(X;18) (SYT-SSX)-positive, regardless of the laboratory involved and the PCR techniques used. PCR is a very sensitive technique. As a consequence, a false-positive result can easily be obtained by means of tissue contamination, even if minimal (i.e., microscopic). The way the tissue is collected from paraffin blocks before being submitted to PCR is crucial. In our opinion, disposable material should be used for each case and PCR-based techniques, especially when they are used for diagnostic purposes, should benefit from quality assurance programs. Along the same line, it should ideally always be possible to verify results from one laboratory by another independent laboratory, using the same material but not necessarily the same technique, as shown in the current study. Real-time PCR, which appeals to strict automation techniques (9), also could improve the quality of the results by decreasing the risk of contamination.
References
Hiraga H, Nojima T, Abe S, Sawa H, Yamashiro K, Yamawaki S, et al. Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol 1998; 7: 102–110.
Willeke F, Mechtersheimer G, Schwarzbach M, Weitz J, Zimmer D, Lehnert T, et al. Detection of SYT-SSX1/2 fusion transcripts by reverse transcriptase-polymerase chain reaction (RT-PCR) is a valuable diagnostic tool in synovial sarcoma. Eur J Cancer 1998; 34: 2087–2093.
Van de Rijn M, Barr FG, Collins MH, Xiong QB, Fisher C . Absence of SYT-SSX fusion products in soft tissue tumors other than synovial sarcoma. Am J Clin Pathol 1999; 112: 43–49.
Guillou L, Coindre JM, Gallagher G, Terrier PH, Gebhard S, de Saint-Aubain Somerhausen N, et al. Detection of the synovial sarcoma translocation t(X;18) (SYT-SSX) in paraffin-embedded tissues using reverse transcriptase-polymerase chain reaction: a reliable and powerful diagnostic tool for pathologists. A molecular analysis of 221 mesenchymal tumors fixed in different fixatives. Hum Pathol 2001; 32: 105–112.
O'Sullivan MJ, Kyriakos M, Zhu X, Wick MR, Swanson PE, Dehner LP, et al. Malignant peripheral nerve sheath tumors with t(X;18). A pathologic and molecular genetic study. Mod Pathol 2000; 13: 1253–1263.
Ladanyi M, Woodruff JM, Scheithauer BW, Bridge JA, Barr FG, Goldblum JR, et al. Letter to the Editor. Mod Pathol 2000; 14: 733–737.
Hostein I, Menard A, Bui BN, Lussan C, Wafflart J, Delattre O, et al. Molecular detection of the synovial sarcoma translocation t(X;18) by real-time PCR in paraffin-embedded material. Diagn Mol Pathol (in press).
Chomczynski P, Sacchi N . Single step method of RNA isolation by acide guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156–159.
Peter M, Gilbert E, Delatre O . A multiplex real-time PCR assay for the detection of gene fusions observed in solid tumors. Lab Invest 2001; 81: 905–912.
Pfeifer JD, Hill DA, O'Sullivan MJ, Dehner LP . Diagnostic gold standard for soft tissue tumours: morphology or molecular genetics? Histopathology 2000; 37: 485–500.
Acknowledgements
This work was supported by the Ligue Nationale Contre le Cancer, Committees of Gironde and Charente-Maritime.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Coindre, JM., Hostein, I., Benhattar, J. et al. Malignant Peripheral Nerve Sheath Tumors are t(X;18)-Negative Sarcomas. Molecular Analysis of 25 Cases Occurring in Neurofibromatosis Type 1 Patients, Using Two Different RT-PCR-Based Methods of Detection. Mod Pathol 15, 589–592 (2002). https://doi.org/10.1038/modpathol.3880570
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3880570
Keywords
This article is cited by
-
Growth-associated protein 43 in differentiating peripheral nerve sheath tumors from other non-neural spindle cell neoplasms
Modern Pathology (2014)
-
TLE1 expression is not specific for synovial sarcoma: a whole section study of 163 soft tissue and bone neoplasms
Modern Pathology (2009)
-
SYT-SSX fusion is absent in sarcomatoid mesothelioma allowing its distinction from synovial sarcoma of the pleura
Modern Pathology (2007)
-
HMGA proteins in malignant peripheral nerve sheath tumor and synovial sarcoma: preferential expression of HMGA2 in malignant peripheral nerve sheath tumor
Modern Pathology (2005)
-
Morphologische Variabilit�t des synovialen Sarkoms im Kindesalter
Der Pathologe (2005)