Abstract
Chromosomal rearrangements involving chromosome bands 12q13–15 are very frequent findings in benign solid tumors, and recently, the primary molecular target for these aberrations was identified as the gene HMGIC. However, mutations in this gene have also been observed in nonneoplastic tissues. In a previous study, we reported breakpoints within HMGIC of synovia affected by osteoarthritis (OA) in two cases with 12q15 aberrations. To analyze further the role of HMGIC in this disease, we have performed cytogenetic, fluorescent in situ hybridization (FISH), RNA, and protein expression analyses on synovial samples from patients with OA and individuals without signs of the disorder. Cytogenetic analysis of short-term cultured cells revealed clonal 12q13–15 aberrations in 2/36 cases of OA synovia and no rearrangement in any of the five controls. With FISH analysis, it was shown that the chromosomal breakpoints in the two aberrant cases were located outside the HMGIC locus. In contrast, at RNA and protein expression analyses, OA-affected as well as normal synovia displayed transcription and translation of the gene. We also analyzed whether immunoreactivity for HMGIC was associated with the proliferation-specific antigen Ki-67, but no correlation between the staining patterns of these proteins was observed. From the results of the present study, it is evident that expression of HMGIC cannot simply be considered a sign of neoplasia or an effect of proliferation.
Similar content being viewed by others
INTRODUCTION
Osteoarthritis (OA) is a nonneoplastic disorder primarily affecting the joint cartilage but also involving the subchondral bone and surrounding synovia (1). Structural changes of the cartilage include fibrillation, ulceration, and cracking. The joint is slowly depleted of cartilage, and sometimes, with advancing disease, the underlying bone is exposed. In the subchondral bone, new bone formation may occur, as well as at the joint margins, where bone proliferation forms so-called osteophytes. The surrounding synovia becomes hyperplastic and shows varying degrees of inflammation. The etiology of OA is not known. However, we have previously described a high incidence of clonal chromosomal alterations in cartilage, osteophytes, and synovia from OA-affected joints (2, 3). Numerical changes are characteristic, with gain of chromosome 7 (trisomy 7), followed by trisomy 5, as the most prominent findings. Also, losses of the sex chromosomes, as well as trisomy 12, are often observed. The only recurrent structural rearrangements detected so far are aberrations involving chromosome bands 12q13–15.
Chromosomal rearrangements involving chromosome bands 12q13–15 are very frequent findings in benign solid tumors including lipomas, uterine leiomyomas, pleomorphic adenomas of the salivary glands, endometrial polyps, and pulmonary chondroid hamartomas (4). Recently, HMGIC was identified to be the target gene affected by these chromosomal aberrations (5, 6). HMGIC belongs to a protein family with two other members, HMGI and HMGY, which are produced by alternative splicing from a gene located in chromosome band 6p21 (7). The proteins are nonhistone components of the chromatin and are characterized by three N-terminal DNA-binding regions and a highly acidic tail in the C-terminal part (8). The DNA-binding regions preferentially bind AT-rich sequences in the minor groove of the chromatin. Several studies indicate that the HMGI family proteins function as general transcription regulatory proteins, which are involved in the organization of the chromatin and the correct assembly and stabilization of other transcription factors at promoter regions (9).
It has been suggested that HMGIC has an important role in the regulation of cell proliferation. The gene is highly expressed in rapidly dividing tissues, like the developing embryo and malignant neoplasias, but in normal human tissues of the adult, HMGIC expression has only been detected in lung and kidney (10, 11). Further support for the regulatory impact of HMGIC in proliferation comes from studies showing that a complete deletion of the gene causes a 40% reduction in body weight in mice (pygmy phenotype) and adipocyte hypoplasia (12, 13). In contrast, Battista et al. (14) showed that mice lacking part of the gene encoding the C-terminal acidic tail have a giant phenotype and develop lipomatosis, indicating that the transforming effect of HMGIC mutations is due to truncation of the protein after the three DNA-binding domains. Further evidence for this notion comes from a study by Fedele et al. (15) demonstrating that only expression of HMGIC lacking the C-terminal part, either in the form of a truncated or fusion protein, can transform murine NIH3T3 cells.
Previously, we showed by fluorescent in situ hybridization (FISH) that HMGIC was the molecular target in two OA cases with translocations in this chromosomal region (16), indicating that rearrangements of this gene are involved in the pathogenesis of OA. In the present study, we analyzed further the involvement of HMGIC in OA. Cytogenetic analysis was used to search for additional cases with rearrangements of 12q13–15 in OA-affected and normal synovia, and FISH was used to establish whether or not HMGIC was involved in such rearrangements. Moreover, we analyzed the expression of HMGIC on the RNA and protein levels in OA-affected and normal synovia.
MATERIALS AND METHODS
Patient Material
Thirty-six synovial samples were retrieved from OA patients undergoing joint replacement, and normal synovial samples were obtained from five patients being operated on for fractures of the femoral neck. All samples derived from either the knee or the hip joint. At the time of surgery, the OA patients and the control individuals were aged 57–84 and 18–90 years, respectively. From each synovial biopsy, one portion was used for short-term culturing followed by cytogenetic and FISH analyses. Another portion was alternatively used for RNA extraction and RT-PCR analysis, or paraffin-embedding and protein expression analysis. The samples for RNA isolation were stored at −80°C until the extraction was performed.
Cytogenetic Analysis
The biopsy product was minced into small pieces using scalpels and subsequently subjected to enzymatic treatment with collagenase II (1000 U/mL; Worthington Biochemical Corporation, Lakewood, NJ) at 37°C for 4 hour. The cell suspensions were spun down, and the cell pellet was resuspended in DMEM/F-12 medium supplemented with 17% fetal calf serum and antibiotics and was plated on chamber slides. After 3–10 days in culture, the cells were automatically harvested using a Tecan©. The harvested chamber slides were dried at 60°C overnight, followed by 2 × SSC treatment at 65°C for 2.5 hours.
The chromosomes were banded using Wright’s stain. For every case 50–100 metaphase cells were analyzed. The clonality criteria and the descriptions of the karyotypes followed the recommendations of the International System for Human Cytogenetic Nomenclature (17).
FISH Analysis
The following probes were used for detection of 12q-rearrangements: the HMGIC-specific cosmids 142H1 and 27E12 (kindly provided by Dr. Schoenmakers, Leuven, Belgium); the YACs 804B7 and 965g6, mapping to chromosome segments 12q24 and 12p11, respectively (kindly provided by CEPH); as well as a 12p-arm–specific chromosome–painting probe (pcp; [18]). The probes were labeled with either digoxigenin–16-dUTP (Boehringer Mannheim, Mannheim, Germany; 142H1, 804B7) or biotin-16-dUTP (Boehringer Mannheim; 27E12, 965g6, pcp12) by using the Megaprime DNA labeling kit from Amersham Pharmacia Biotech (Little Chalfont, UK).
The chamber slides used for the FISH hybridizations were processed in the same way as for the cytogenetic analysis. Before the denaturation, the slides were treated with pepsin (10 mg/mL, pH 2; Serva, Heidelberg, Germany) at 37°C for 10 minutes, washed in 1 × phosphate buffered saline (PBS) twice for 3 minutes each, postfixed in 1% formaldehyde for 10 minutes, washed in 1 × PBS twice for 3 minutes each, and finally dehydrated in an ethanol series (70%, 85%, and 96%). Thereafter, the slides were overlaid with denaturation solution (70% formaldehyde, 2 × SSC) and denatured on a heating plate at 72°C for 2 minutes, followed by dehydration in an ethanol series. The probes were denatured at 70°C for 10 minutes and subsequently prehybridized at 37°C for 40 minutes before application on the denatured chamber slides. The hybridizations were performed in a humidified chamber at 37°C overnight.
Stringent washes were performed in 0.4 × SSC at 70°C for 2 minutes. Then, the slides were rinsed in TNT (0.1 m Tris-HCl, pH 7.5; 0.15 m NaCl; 0.05% Tween 20) for 3 minutes, incubated with TNB blocking solution (Boehringer Mannheim) at room temperature for 30 minutes, and rinsed in TNT for another 3 minutes. For detection of the probes, the slides were incubated with Cy3-conjugated avidin (1:1000; Amersham Pharmacia Biotech) and fluorescent-conjugated anti-digoxigenin antibodies (1:100; Boehringer Mannheim) at room temperature for 30 minutes. The slides were finally washed in TNT three times for 3 minutes each, followed by dehydration in an ethanol series. The chromosomes were counterstained with DAPI (4′,6-Diamidine-2′-phenylindole-dihydrochloride; Boehringer Mannheim), and the hybridization signals were detected in an Axioscop fluorescence microscope (Zeiss, Jena, Germany) and analyzed with CytoVision Ultra system software (Applied Imaging, Newcastle, UK). For every case, approximately 100 metaphase cells were analyzed.
RNA Extraction, cDNA Synthesis, and Expression PCR
mRNA was isolated from synovial biopsies according to the protocol provided from Dynal (Oslo, Norway), and for the subsequent synthesis of single-stranded cDNA, the following procedure was performed: mRNA was incubated with 10 ng of random hexamers in a 10-μL volume at 65°C for 5 minutes. Then, a solution with the following concentrations was added to the mRNA: first-strand buffer (1 ×; Gibco BRL, Täby, Sweden), dithiothreitol (0.1 m; Gibco BRL), RNA Guard (3.14 U/μL; Amersham Pharmacia Biotech), dNTP (40 mm), and M-MLV reverse transcriptase (2 U/μL; Gibco BRL), and this reaction mixture was incubated at 37°C for 1 hour. Another 20 U of M-MLV reverse transcriptase was added to the reaction mixture, followed by incubation at 42°C for 30 minutes and a final step at 50°C for 10 minutes.
The HMGIC-specific primers used for the expression analysis were GGCCGTTTTTCTCCAGTG for antisense primer, corresponding to nucleotides 1038–1021, and ACTTCAGCCCAGGGACAA for sense primer, corresponding to 848–865 in the cDNA sequence (Genbank accession number U28749). As positive control of RNA integrity, a 626-bp fragment of ACTB was simultaneously amplified using the primers CCTCGCCTTTGCCGATCC and GGATCTTCATCATGAGGTAGTCAGTC (19). For the PCR, 1–6 μL of 1:20 diluted single-stranded cDNA was used as template. As negative and positive controls of HMGIC expression, cDNA template from adipose tissue and cultured amniocytes were used. The PCR reaction of 50 μL contained 50 mm KCl, 10 mm Tris-HCl, pH 8.3, 1.25 mm MgCl2, 0.2 mm of each dNTP, 4 U of Platinum Taq polymerase (Gibco BRL), and 0.8 mm of each primer for HMGIC. The reaction mixture was initially denatured for 5 minutes at 96°C, followed by 10 cycles of a sequence of 30 seconds at 95°C, 30 seconds at 59°C, and 1 minute at 72°C. Thereafter, 0.4 mm of each of the primers specific for ACTB were added at 10°C, followed by another 25 cycles as described. The PCR products were separated on a 1.5% agarose gel, and the intensities were visualized through staining with VistraGreen (1:10,000; Amersham Pharmacia Biotech).
Immunohistochemical Analysis
From routinely processed paraffin-embedded tissue blocks, 5-μm thick sections were cut and immunostained, according to established protocols, using an avidin-biotinylated peroxidase complex (ABC) technique after antigen retrieval (Target Retrieval Solution; DAKO, Glostrup, Denmark). The HMGIC antibodies were raised in rabbit against the recombinant murine HMGIC protein (20). For detection of Ki-67 expression, a monoclonal IgG mouse antibody (MIB1; Immunotech/Coulter, Fullerton, CA) was used. The HMGIC and MIB1 antibodies were used at 1:400 and 1:320 dilutions, respectively. Negative controls were performed by omitting the primary antibody. The proportion of HMGI-C and Ki-67 positive cells (labeling index) was determined by estimating the percentage of positive nuclei after counting 1000 cells from randomly selected areas of the synovium at 400 ×. Only cases in which positive nuclear staining was observed in more than 10% of the cells were scored as positive of HMGIC.
RESULTS
Cytogenetic and FISH analysis
The most common chromosomal aberration detected was trisomy 7, which was present in 34/36 of OA-affected and 3/5 normal cases (Table 1). Trisomy 5 was found in 7/36 OA cases and in 1/5 normal synovial samples. Loss of chromosome Y was observed in 7/12 cases from men with OA and in 2/3 male controls. In 2/36 cases of OA-affected synovia (Cases 1 and 2), clonal rearrangements involving 12q13–15 were detected. No rearrangements in this region were observed among the normal cases.
FISH analysis showed that in none of the two cases with clonal 12q13–15 aberrations was the breakpoint located within the HMGIC gene (data not shown). FISH was also performed on Cases 3–21, but none of these displayed cryptic rearrangements of the gene. In 15/21 cases, nonclonal aberrations involving chromosome 12 were detected (Table 1). Trisomy 12 was the most common finding, and HMGIC was not involved in any of the structural rearrangements observed.
RNA and Protein Expression Analyses
In 10/14 cases of OA synovia and in the two cases of normal synovia, expression of HMGIC was detected at the RNA level (Table 1; Fig. 1). Because some of the cases displayed low expression, the expression experiments were performed several times in order to confirm the positive cases. No attempt to quantify the expression was made.
The picture shows some of the cases analyzed for expression of HMGIC at the RNA level. cDNAs from OA-affected and normal synovial biopsies were subjected to PCR with HMGIC-specific primers, resulting in a 192-bp fragment. As an internal standard, amplification with ACTB-specific primers for a 626-bp fragment was simultaneously performed. Lane 1: Case 26 (OA-affected synovia); Lane 2: Case 24 (OA); Lane 3: Case 25 (OA); Lane 4: Case 29 (OA); Lane 5: Case 34 (OA); Lane 6: Case 40 (normal synovia); Lane 7: Case 41 (normal synovia); Lane 8: normal adipose tissue; Lane 9: cultured amniocytes.
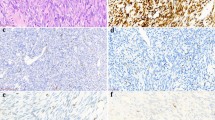
The immunohistochemical analysis showed immunoreactivity localized to the nucleus of the synovial lining cells (Fig. 2). A few stromal cells adjacent to the synovial lining cells also showed nuclear reactivity. The inflammatory cells were negative for HMGIC. Immunoreactivity was identified in 11/21 of the OA-affected cases and in 1/4 of the normal cases (Table 1). There was no obvious correlation between the immunohistochemistry results and the degree of synovitis. However, positive staining was greater in areas featuring multilayering of synovial lining cells and increased cellularity (Fig. 2). The positive nuclei were observed among scattered negative cells, arguing against the possibility of false-positive staining in these experiments.
A, Hematoxylin and eosin section of the synovial biopsy from Case 37 (40 ×). The synovium shows reactive changes with increased cellularity and multilayering of synovial lining cells. The area within the black rectangle is shown after immunostaining with HMGIC-specific antibodies at higher magnification in Panel B (400 ×). The immunoreactivity was localized to the cell nucleus of the synovial lining cells. Negative control was performed by omitting the primary antibody (C).
All cases showed very low proportions of Ki-67–positive cells (<5%). There was no correlation between Ki-67 expression and the degree of positive staining for HMGIC.
DISCUSSION
The cytogenetic investigation presented here revealed that the only recurrent structural rearrangement in synovial short-term cultures from patients with OA affected the chromosomal region 12q13–15, which is in concordance with previous reports of the karyotypic pattern of OA (2, 3). However, in neither of the two aberrant cases identified did the translocation involve the HMGIC gene, nor were any cryptic subchromosomal aberrations of the gene found in any of the cases analyzed. Breakpoints outside the coding region of HMGIC do not, however, exclude the possibility that expression of the gene is affected by the chromosomal alterations. Genomic rearrangements outside HMGIC associated with increased expression have been reported in uterine leiomyomas (11, 21), and it has been suggested that these locus-distant breakpoints might affect up- or downstream regulatory elements of importance for the HMGIC expression. Furthermore, in a subset of lipomas with 12q13–15 rearrangements, the breakpoints have been located more than 10 Mb away from the HMGIC gene, indicating that breakpoints in adjacent genes might give rise to the same cellular phenotype as rearrangements involving HMGIC (22).
However, the results from the present study showed that transcription and translation of HMGIC in synovia from adults are not associated with breakpoints of the 12q13–15 region. Although no mutations of HMGIC were found in any case by FISH analysis, expression of the gene at the RNA level was present in the majority of the OA-affected synovial samples, as well as in the two normal cases analyzed. Also, the immunohistochemical analysis revealed positive staining of the protein in approximately half of the OA-affected synovial samples and in one without signs of the disease. Thus, HMGIC expression in synovia is not specific for neoplastic proliferation, nor OA-associated changes, but that the large proportion of cases was positive indicates that expression of HMGIC might be of importance in other processes of the synovia.
HMGIC expression has not been analyzed previously in synovia or any other joint tissue. Two earlier studies have shown that HMGIC is not expressed in normal adult tissues (11, 23), except for weak expression observed in lung and kidney. The results of the present study indicate that other factors than disruption of the HMGIC locus might upregulate expression of the gene in nonneoplastic mature cells. Microenvironmental factors might be of importance, affecting a limited part of the cell population. Indeed, the rather weak expression at the RNA level, and furthermore, the finding of HMGIC-positive cells scattered among negative cells, indicate that only a fraction of the cell population produces the protein in synovia.
Because it has been proposed that HMGIC is involved in the regulation of cellular proliferation, we analyzed whether the staining pattern for the proliferation-specific Ki-67 antigen was associated with the degree of positive staining for the HMGIC protein in the tissue samples. However, similar to what has been shown in other studies (24), the proliferative activity observed was very restricted, and there was no correlation between HMGIC and Ki-67. Our finding of HMGIC expression in an adult differentiated tissue type with low-proliferating activity complicates the interpretation of the role of HMGIC in neoplasia and indicates the need for additional studies to better understand its possible function in mature cell types.
Accession codes
References
Brandt KD, Mankin HJ . Pathogenesis of osteoarthritis. In: Kelley WN, Harris ED, Ruddy S, Sledge CB, editors. Textbook of rheumatology. 4th ed. Philadelphia: Saunders; 1993. pp. 1355–1373.
Mertens F, Pålsson E, Lindstrand A, Toksvig-Larsen S, Knuutila S, Larramendy ML, et al. Evidence of somatic mutations in osteoarthritis. Hum Genet 1996; 98: 651–656.
Broberg K, Limon J, Pålsson E, Lindstrand A, Toksvig-Larsen S, Mandahl N, et al. Clonal chromosome aberrations are present in vivo in synovia and osteophytes from patients with osteoarthritis. Hum Genet 1997; 101: 295–298.
Mitelman F, editor. Catalog of chromosome aberrations in cancer. ’98, CD-ROM version 1. New York: Wiley-Liss; 1998.
Ashar HR, Fejzo MS, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, et al. Disruption of the architectural factor HMGI-C: DNA-binding AT motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 1995; 82: 57–65.
Schoenmakers EPM, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJM . Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet 1995; 10: 436–444.
Johnson KR, Lehn DA, Elton TS, Barr PJ, Reeves R . Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y). J Biol Chem 1988; 263: 18338–18342.
Bustin M, Reeves R . High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol 1996; 54: 35–100.
Carey M . The enhanceosome and transcriptional synergy. Cell 1998; 92: 5–8.
Patel UA, Bandeira A, Manfioletti G, Giancotti V, Chay KY, Crane-Robinson C . Expression and cDNA cloning of human HMGI-C phosphoprotein. Biochem Biophys Res Commun 1994; 201: 63–70.
Gattas GJF, Quade BJ, Novak RA, Morton CC . HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer 1999; 25: 316–322.
Xiang X, Benson KF, Chada K . Minimouse: disruption of the pygmy locus in a transgenic insertional mutant. Science 1990; 247: 967–969.
Zhou X, Benson KF, Chada K . Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995; 376: 771–774.
Battista S, Fidanza V, Fedele M, Klein-Szanto AJ, Outwater E, Brunner H, et al. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res 1999; 59: 4793–4797.
Fedele M, Berliengieri MT, Scala S, Chiariotti L, Viglietto G, Rippel V, et al. Truncated and chimeric HMGI-C genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene 1998; 17: 413–418.
Broberg K, Höglund M, Limon J, Lindstrand A, Toksvig-Larsen S, Mandahl N, et al. Rearrangement of the neoplasia-associated gene HMGIC in synovia from patients with osteoarthritis. Genes Chromosomes Cancer 1999; 24: 278–282.
Mitelman F, editor. ISCN. An international system for human cytogenetic nomenclature. Basel: Karger; 1995.
Zhang J, Marynen P, Devriendt K, Fryns J, Van den Berghe H, Cassiman J . Molecular analysis of the isochromosome 12P in the Pallister-Killian syndrome. Construction of a mouse-human hybrid cell line containing an i(12p) as the sole human chromosome. Hum Genet 1989; 83: 359–363.
Raff T, van der Giet M, Endemann D, Wiederholt T, Paul M . Design and testing of β-actin primers for RT-PCR that do not co-amplify processed pseudogenes. Biotechniques 1997; 23: 456–460.
Tallini G, Dal Cin P, Rhoden KJ, Chiapetta G, Manfioletti G, Giancotti V, et al. Expression of HMGI-C and HMGI(Y) in ordinary lipoma and atypical lipomatous tumors. Am J Pathol 1997; 151: 37–43.
Schoenberg Fejzo MS, Ashar HR, Krauter KS, Powell WL, Rein MS, Weremowicz S, et al. Translocation breakpoints upstream of the HMGIC gene in uterine leiomyomata suggest dysregulation of this gene by a mechanism different from that in lipomas. Genes Chromosomes Cancer 1996; 17: 1–6.
Merscher S, Marondel Y, Pedetour F, Gaudray P, Kucherlapati R, Turc-Carel C . Identification of new translocation breakpoints at 12q13 in lipomas. Genomics 1997; 46: 70–77.
Rogalla P, Drechsler K, Frey G, Hennig Y, Helmke B, Bonk U, et al. HMGI-C expression patterns in human tissues. Am J Pathol 1996; 149: 775–779.
Lalor PA, Mapp PI, Hall PA, Revell PA . Proliferative activity of cells in the synovium as demonstrated by a monoclonal antibody, Ki67. Rheumatol Int 1987; 7: 183–186.
Ahlbäck S . Osteoarthritis of the knee. A radiographic investigation. Acta Radiol Suppl 1968; 277: 1–72.
Boegård T, Jonsson K . Radiography in osteoarthritis of the knee. Skeletal Radiol 1999; 28: 605–615.
Acknowledgements
This work was supported by the AndersOtto Swärd Foundation, the IngaBritt and Arne Lundberg Research Foundation, the Nilsson-Ehle Foundation, and the Gustav V 80 years Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Broberg, K., Tallini, G., Höglund, M. et al. The Tumor-Associated Gene HMGIC Is Expressed in Normal and Osteoarthritis-Affected Synovia. Mod Pathol 14, 311–317 (2001). https://doi.org/10.1038/modpathol.3880308
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3880308