Abstract
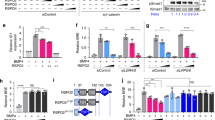
THE TGF-β/activin/BMP cytokine family signals through serine/threonine kinase receptors, but how the receptors transduce the signal is unknown. The Mad (Mothers against decapentaplegic) gene from Drosophila1 and the related Sma genes from Caenorhabditis elegans2 have been genetically implicated in signalling by members of the bone-morphogenetic-protein (BMP) subfamily. We have cloned Smad1, a human homologue of Mad and Sma. Microinjection of Smad1 messenger RNA into Xenopus embryo animal caps mimics the mesoderm-ventralizing effects of BMP4. Smad1 moves into the nucleus in response to BMP4. Smad1 has transcriptional activity when fused to a heterologous DNA-binding domain, and this activity is increased by BMP4 acting through BMP-receptor types I and II. The transactivating activity resides in the conserved carboxy-terminal domain of Smad1 and is disrupted by a nonsense mutation that corresponds to null mutations found in Mad and in the related gene DPC4, a candidate tumour-suppressor gene in human pancreatic cancer3. Additionally, we show that DPC4 contains a transcriptional activation domain. The results suggests that the Smad proteins are a new class of transcription factors that mediate responses to the TGF-β family.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Sekelsky, J. J., Newfeld, S. J., Raftery, L. A., Chartoff, E. H. & Gelbart, W. M. Genetics 139, 1347–1358 (1995).
Savage, C. et al. Proc. natn. Acad. Sci. U.S.A. 93, 790–794 (1996).
Hahn, S. A. et al. Science 271, 350–353 (1996).
Dale, L., Howes, G., Price, B. M. J. & Smith, J. C. Development 115, 573–585 (1992).
Jones, C. M., Lyons, K. M., Lapan, P. M., Wright, C. V. E. & Hogan, B. L. M. Development 115, 639–647 (1992).
Graff, J. M., Thies, R. S., Song, J. J., Celeste, A. J. & Melton, D. A. Cell 79, 169–179 (1994).
Suzuki, A. et al. Proc. natn. Acad. Sci. U.S.A. 91, 10255–10259 (1994).
Harland, R. M., Proc. natn. Acad. Sci. U.S.A. 91, 10243–10246 (1994).
Cárcamo, J. et al. Molec. cell. Biol. 14, 3810–3821 (1994).
Ptashne, M. Nature 335, 683–689 (1988).
Lillie, J. W. & Green, M. R. Nature 338, 39–44 (1989).
Liu, F., Ventura, F., Doody, J. & Massagué, J. Molec. cell. Biol. 15, 3479–3486 (1995).
Rosenzweig, B. L. et al. Proc. natn. Acad. Sci. U.S.A. 92, 7632–7636 (1995).
Lennon, G. G., Auffray, C., Polymeropoulos, M. & Soares, M. B. Genomics (in the press).
Rupp, R. A. & Weintraub, H. Cell 65, 927–937 (1991).
Wilson, P. A. & Melton, D. A. Curr. Biol. 4, 676–686 (1994).
Lamb, T. M. et al. Science 262, 713–718 (1993).
Sadowski, I. & Ptashne, M. Nucleic Acids Res. 17, 7539 (1989).
Wrana, J. L., Attisano, L., Wieser, R., Ventura, F. & Massagué, J. Nature 370, 341–347 (1994).
Gorman, C. M., Moffat, L. F. & Howard, B. H. Molec. cell. Biol. 2, 1044–1051 (1982).
Liu, F. & Green, M. R. Cell 61, 1217–1224 (1990).
Hoodless, P. A. et al. Cell 85, 489–500 (1996).
Graff, J. M., Bansal, A. & Melton, D. A. Cell 85, 479–487 (1996).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Liu, F., Hata, A., Baker, J. et al. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature 381, 620–623 (1996). https://doi.org/10.1038/381620a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/381620a0
This article is cited by
-
In Silico Analysis: Genome-Wide Identification, Characterization and Evolutionary Adaptations of Bone Morphogenetic Protein (BMP) Gene Family in Homo sapiens
Molecular Biotechnology (2023)
-
BMP3 is a novel locus involved in the causality of ocular coloboma
Human Genetics (2022)
-
USP7 facilitates SMAD3 autoregulation to repress cancer progression in p53-deficient lung cancer
Cell Death & Disease (2021)
-
Endothelial struts enable the generation of large lumenized blood vessels de novo
Nature Cell Biology (2021)
-
Bone morphogenetic protein 7 promotes resistance to immunotherapy
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.