Abstract
Loss of chromosome 9p has been implicated in the progression of renal cell carcinoma. We evaluated the clinical utility of fluorescence in situ hybridization analysis of loss of chromosome 9p in 73 patients with clear cell renal cell carcinomas with varied stage, size, grade, necrosis (SSIGN) scores. Loss of chromosome 9p was observed in 13 tumors (18%). The 5-year cancer-specific survival of patients without loss of chromosome 9p was 88% and was 43% in those with loss of chromosome 9p (P<0.001). Local extension of the primary tumor according to the 2002 TNM staging system, lymph node involvement, the presence of distant metastases, and the SSIGN score were the other variables that predicted cancer-specific survival in univariate analysis. Loss of chromosome 9p was an independent prognostic factor in multivariate analysis. Our data indicate that the detection of chromosome 9p loss by fluorescence in situ hybridization analysis of clear cell renal cell carcinoma adds prognostic information beyond the pathological factors included in the current predictive models for renal cell carcinoma, such as SSIGN score.
Similar content being viewed by others
Introduction
Several models have been recently developed to better stratify patients with renal cell carcinomas into different prognostic categories.1, 2, 3, 4 The Kattan nomogram includes histologic type, tumor size, TNM stage, and clinical presentation to estimate the probability of 5-year recurrence-free survival.3 The UISS model includes TNM stage, Eastern Cooperative Oncology Group (ECOG) performance status, and Fuhrman nuclear grade.2, 5, 6 The stage, size, grade, necrosis (SSIGN) score developed by Frank et al4 includes tumor stage, tumor size, tumor grade, and necrosis. The SSIGN score has recently been validated in a European study of patients who were surgically treated for clear cell renal cell carcinoma.7 Reliable prognostic indicators that can distinguish among different prognostic groups are important in predicting outcome and in qualifying subjects for clinical trials of new therapies. Loss of chromosome 9p has been found to be associated with worse prognosis in patients with clear cell renal cell carcinoma.8 This study was undertaken to assess whether loss of chromosome 9p is an independent predictor of cancer-specific survival in patients with clear cell renal cell carcinoma with varied SSIGN scores.
Materials and methods
Patients
Seventy-three patients were selected from the Verona Renal Cell Carcinoma Database. The patients were stratified into five groups (SSIGN scores 0–2, 3 and 4, 5 and 6, 7–9, and 10 or greater) as reported previously.7 The patients for this study were selected randomly from among those with in the database with clear cell renal cell carcinoma and required SSIGN scores.
The clinical characteristics analyzed included patients' age, gender, ECOG performance status, symptoms at presentation, and surgical procedure. Their symptoms were classified according to Patard's system.9 The pathological features that were studied included tumor size, 2002 primary tumor T classification, regional lymph node involvement, distant metastases, Fuhrman's nuclear grade, and coagulative tumor necrosis. As reported previously, all slides were reviewed.10 Coagulative tumor necrosis was identified histologically by the presence of homogeneous clusters and sheets of degenerating and dead cells. Degenerative changes such as hyalinization, hemorrhage, and fibrosis were not considered tumor necrosis.4
Tissue Samples
The tissue microarrays were constructed using a tissue-arraying instrument (Beecher Instruments, Hackensack, NJ, USA). Tissue cylinders with diameters of 0.6 mm were punched from each donor paraffin block in targeted areas corresponding to the previously demarcated areas on the parallel hematoxylin- and eosin-stained slide. These tissue cores were then deposited into a recipient, ‘master’ paraffin block. The punches were placed 1 mm apart on the x axis and 1.5 mm apart on the y axis. Each microarray block contained a maximum of 79 punches.
Sections 5-μm thick were cut from the master block, stained with hematoxylin and eosin, and reviewed to ensure the presence of morphologically similar cores of clear cell renal cell carcinomas. Morphologic features of each core were confirmed by reviewing the corresponding whole-tissue sections stained with hematoxylin and eosin. Sections were then used for in situ hybridization.
At least three neoplastic cores were available for neoplastic tissue and at least two cores from normal renal parenchyma adjacent to the carcinoma were included from each case. Overall, 225 cores of clear cell renal cell carcinoma and 150 samples of normal renal parenchyma adjacent to the carcinomas were included in five tissue microarrays.
Fluorescence In Situ Hybridization
We performed interphase cytogenetic fluorescence in situ hybridization (FISH) analysis using a telomere-specific probe (115 kb) for the chromosome 9p telomere (SpectrumOrange LSI, Abbott) and a centromeric (α-satellite DNA) probe for the region 9p11–q11. FISH was also performed on whole-tissue sections for 17 of the tumors.
From the whole-tissue sections and tissue microarrays, 5 μm sections were cut from paraffin-embedded blocks. The paraffin was removed from the sections with two 10-min washes with xylene. After hydrating in 100, 85, and 70% ethanol solutions (10 min), rinsing in distilled water (10 min), and twice in phosphate-buffered solution (pH 7, 10 min each), the slides were fixed in methanol-acetic acid 3:1 for 10 min and air-dried. Next, the sections were treated in a 2 × standard saline citrate solution for 15 min at 37°C, and then dehydrated in consecutive 70, 85, and 100% ethanol solutions for 1 min each and then dried. Next, the sections were bathed in 0.1 mM citric acid (pH 6) solution at 85°C for 1 h. Then they were again dehydrated in a series of ethanol solutions and dried. The tissue was digested by applying 0.75 ml of pepsin (Sigma, St Louis, MO, USA) solution (4 mg/ml in 0.9% NaCl, pH 1.5) to each slide and incubating them in a humidified box for 30 min at 37°C. Next, the slides were rinsed with distilled water for a few seconds, dehydrated again in graded ethanol solutions, and dried. Centromeric probes for chromosomes 9 and locus-specific probes for 9p (Vysis, Downers Grove, IL, USA) were used. Each probe was diluted 1:100 in tDenHyb1 buffer (Insitus, Albuquerque, NM, USA). Ten microliters of diluted probe was applied to each slide and cover slips were placed over the slides. Denaturation was achieved by incubating the slides at 80°C for 10 min in a humidified box, and then hybridization was carried out at 37°C for 16 h. The cover slips were later removed and the slides were immersed at room temperature in 0.5 XSSC for 2 min, in 50% formamide/1 XSSC for 5 min, and in 2 XSSC for 2 min. The slides were air-dried and counterstained with 10 μl DAPI/Antifade (DAPI in Fluorguard, 0.5 μg/ml; Insitus).
The slides were examined using an Axioplan microscope (Zeiss, Germany) with filters for SpectrumOrange (Tel 9p, Vysis-Abbott), SpectrumGreen (centromeric probe 9, Vysis-Abbott), and the UV Filter for the DAPI nuclear counterstain. The signals were recorded with a CCD camera (Axiocam HRm).
Fluorescent signals were evaluated as reported previously.11, 12 Signals from 100–200 neoplastic nuclei were counted. Scoring of the microarray was performed as follows: the results for individual punches were compared with their replicas and a consensus score was recorded for each tumor. The criteria for loss of chromosome 9p were described previously.13, 14 Briefly, chromosomal loss was defined as a percentage of nuclei with single signals greater than the mean number of nuclei with single signals plus four times the standard deviation found in the samples of normal renal parenchyma. Thus, for centromeric chromosomes 9 and telomeric 9p, percentages of single signals greater than 30 and 31%, respectively, were considered to indicate loss.
Statistical Analysis
Cancer-specific survival was estimated using the Kaplan–Meier method. Follow-up was calculated from the date of surgery to the date of death or last clinical evaluation. The cause of death was determined from correspondence with the family of the patient or local physician, or the death certificate. Association of the clinical and pathological features studied with death from renal cell carcinoma were evaluated using the log rank test and the Cox proportional hazards regression models.
Considering the potential collinearity between single pathological variables and SSIGN score, we generated two different multivariate models. The first one included all variables that were significant in univariate analysis, and the second one included SSIGN score and all other clinical or pathological variables not included in the SSIGN algorithm that were significant in univariate analysis. Statistical analyses were performed using the SPSS software package, 14.0 version with P<0.05 considered statistically significant.
Results
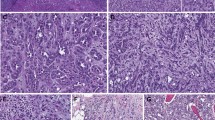
Table 1 summarizes the pathological characteristics of the patients. The median age was 65 years (range 54–71). Fifty-five (75%) were males and 18 (25%) females. The ECOG performance status was ‘0’ for 70 (96%) patients, ‘1’ for 2 patients (3%), and ‘3’ for 1 patient (1%). All tumors were clear cell renal cell carcinoma (Figure 1). Thirty-five (48%) tumors were incidentally detected and 38 (52%) were symptomatic at presentation. The treatment of the primary tumor was radical nephrectomy in 66 (90%) patients and partial nephrectomy in 7 (10%). The median size of the tumors was 6 cm. The pathologic stages of the carcinomas were: pT1a, 14 patients (19%); pT1b, 21 (29%); pT2, 10 (14%); pT3a, 10 (14%); pT3b, 17 (23%); and pT4, 1 (1%) case. Lymph node metastases were present in five patients (7%) and absent in 68 (93%) cases. Remote metastases were absent in 66 patients (90%) and present in 7 (10%). The Fuhrman nuclear grades were 1–4 in 3 (4%), 24 (33%), 34 (47%), and 12 (16%) patients, respectively. Necrosis was absent in 61 carcinomas (84%) and present in 12 (16%). SSIGN scores were distributed as follows: 0–2 in 21 (29%), 3–4 in 24 (33%), 5–6 in 13 (18%), 7–9 in 12 (16%), and 10 or >10 in 3 (4%).
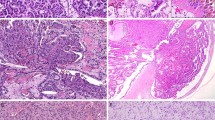
Loss of chromosome 9p was observed in 13 carcinomas (18%). These tumors showed single signals in most nuclei (Figure 2), ranging from 44 to 66% (mean 54%), on whole-tissue sections and from 39 to 69% (mean 55%) in the three tissue array cores. Overall, the concordance between the whole sections and the tissue microarrays was high (concordance index 0.80).
The median follow-up was 45 months (IQR 24–78). At follow-up, 56 (77%) patients were alive and disease-free, whereas 16 (22%) died of renal cell carcinoma and 1 (1%) died of other causes. The probability of 5-year cancer-specific survival was 88% in patients without loss of chromosome 9p and 43% in those with loss of chromosome 9p (log rank P-value <0.001; Figure 3). Local extension of the primary carcinoma, lymph node metastasis, distant metastasis, and SSIGN score were significant predictors of cancer-specific survival in the univariate analysis (Table 2). Loss of chromosome 9p was an independent predictive factor in the multivariate analysis, both in the first model including local tumor extension (pT), lymph node metastasis (N), distant metastasis (M), and in the second one including only SSIGN score (Table 3).
Discussion
In this study, we found that loss of chromosome 9p is a prognostic factor for patients with clear cell renal cell carcinoma, which is independent of SSIGN score and other important prognostic factors. Loss of chromosome 9p was observed in 18% of clear cell renal cell carcinomas. The 5-year cancer-specific survival was 88% in patients without loss of chromosome 9p, compared to 43% in those with loss of chromosome 9p. Models to predict outcome have recently been developed for patients with renal cell carcinomas.1, 2, 3, 4 These systems can be used to counsel patients, determine the need for adjuvant therapy, stratify patients for clinical trials, and develop appropriate postoperative surveillance programs that are tailored to a patient's risk for cancer progression. Loss of chromosome 9p adds new prognostic information beyond the factors included in the current models.2, 4 The significant difference in cancer-specific survival between patients with tumors and without 9p losses suggests that a tumor suppressor gene on 9p may be involved in tumor progression.15, 16, 17, 18 Loss of chromosome 9p was found in 12–36% of clear cell renal cell carcinomas by microsatellite analyses and comparative genomic hybridization.19, 20, 21 Bissig et al22 found losses of chromosome 9p more frequently in metastases but not in the corresponding primary tumors. Cairns et al17 reported deletions in 14 of 42 primary clear cell renal cell carcinomas (33%). Other groups reported 9p loss of heterozygosity in clear cell renal cell carcinomas.16, 19, 23, 24, 25 Presti et al26 suggested that loss of heterozygosity on chromosomes 8p or 9p may be prognostically significant in patients with locally advanced renal cell carcinoma. Schraml et al18 demonstrated an association between loss of heterozygosity on chromosome 9p (24% of cases), using highly polymorphic microsatellite markers, and survival of patient for short duration. Kinoshita et al27 detected a hemi- or homozygous deletion in 9p21–22 region in 3 out of 5 renal cell carcinoma metastases. Gunawan et al28 found association between loss of chromosome 9p and distant metastasis at diagnosis.
These studies suggest that chromosome 9p may be implicated in cancer progression in patients with renal cell carcinoma. A tumor suppressor gene proximal to the CDKN2A locus and protein p16 have been suggested to play a role in clear cell renal cell carcinomas development.18
References
Sorbellini M, Kattan MW, Snyder ME, et al. A postoperative prognostic nomogram predicting recurrence for patients with conventional clear cell renal cell carcinoma. J Urol 2005;173:48–51.
Zisman A, Pantuck AJ, Dorey F, et al. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol 2001;19:1649–1657.
Kattan MW, Reuter V, Motzer RJ, et al. A postoperative prognostic nomogram for renal cell carcinoma. J Urol 2001;166:63–67.
Frank I, Blute ML, Cheville JC, et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol 2002;168:2395–2400.
Zisman A, Pantuck AJ, Wieder J, et al. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J Clin Oncol 2002;20:4559–4566.
Zisman A, Pantuck AJ, Figlin RA, et al. Validation of the ucla integrated staging system for patients with renal cell carcinoma. J Clin Oncol 2001;19:3792–3793.
Ficarra V, Martignoni G, Lohse C, et al. External validation of the mayo clinic stage, size, grade and necrosis (SSIGN) score to predict cancer specific survival using a European series of conventional renal cell carcinoma. J Urol 2006;175:1235–1239.
Jones TD, Eble JN, Cheng L . Application of molecular diagnostic techniques to renal epithelial neoplasms. Clin Lab Med 2005;25:279–303.
Patard JJ, Leray E, Rodriguez A, et al. Correlation between symptom graduation, tumor characteristics and survival in renal cell carcinoma. Eur Urol 2003;44:226–232.
Ficarra V, Martignoni G, Maffei N, et al. Original and reviewed nuclear grading according to the Fuhrman system: a multivariate analysis of 388 patients with conventional renal cell carcinoma. Cancer 2005;103:68–75.
Cossu-Rocca P, Eble JN, Delahunt B, et al. Renal mucinous tubular and spindle carcinoma lacks the gains of chromosomes 7 and 17 and losses of chromosome Y that are prevalent in papillary renal cell carcinoma. Mod Pathol 2006;19:488–493.
Brunelli M, Eble JN, Zhang S, et al. Metanephric adenoma lacks the gains of chromosomes 7 and 17 and loss of Y that are typical of papillary renal cell carcinoma and papillary adenoma. Mod Pathol 2003;16:1060–1063.
Brunelli M, Eble JN, Zhang S, et al. Eosinophilic and classic chromophobe renal cell carcinomas have similar frequent losses of multiple chromosomes from among chromosomes 1, 2, 6, 10, and 17, and this pattern of genetic abnormality is not present in renal oncocytoma. Mod Pathol 2005;18:161–169.
Cossu-Rocca P, Eble JN, Zhang S, et al. Acquired cystic disease-associated renal tumors: an immunohistochemical and fluorescence in situ hybridization study. Mod Pathol 2006;19:780–787.
Junker K, Weirich G, Amin MB, et al. Genetic subtyping of renal cell carcinoma by comparative genomic hybridization. Recent Results Cancer Res 2003;162:169–175.
Schullerus D, Herbers J, Chudek J, et al. Loss of heterozygosity at chromosomes 8p, 9p, and 14q is associated with stage and grade of non-papillary renal cell carcinomas. J Pathol 1997;183:151–155.
Cairns P, Tokino K, Eby Y, et al. Localization of tumor suppressor loci on chromosome 9 in primary human renal cell carcinomas. Cancer Res 1995;55:224–227.
Schraml P, Struckmann K, Bednar R, et al. CDKNA2A mutation analysis, protein expression, and deletion mapping of chromosome 9p in conventional clear-cell renal carcinomas: evidence for a second tumor suppressor gene proximal to CDKN2A. Am J Pathol 2001;158:593–601.
Moch H, Presti Jr JC, Sauter G, et al. Genetic aberrations detected by comparative genomic hybridization are associated with clinical outcome in renal cell carcinoma. Cancer Res 1996;56:27–30.
Jiang F, Desper R, Papadimitriou CH, et al. Construction of evolutionary tree models for renal cell carcinoma from comparative genomic hybridization data. Cancer Res 2000;60:6503–6509.
Gronwald J, Storkel S, Holtgreve-Grez H, et al. Comparison of DNA gains and losses in primary renal clear cell carcinomas and metastatic sites: importance of 1q and 3p copy number changes in metastatic events. Cancer Res 1997;57:481–487.
Bissig H, Richter J, Desper R, et al. Evaluation of the clonal relationship between primary and metastatic renal cell carcinoma by comparative genomic hybridization. Am J Pathol 1999;155:267–274.
Bugert P, Kovacs G . Molecular differential diagnosis of renal cell carcinomas by microsatellite analysis. Am J Pathol 1996;149:2081–2088.
Thrash-Bingham CA, Salazar H, Freed JJ, et al. Genomic alterations and instabilities in renal cell carcinomas and their relationship to tumor pathology. Cancer Res 1995;55:6189–6195.
Grady B, Goharderakhshan R, Chang J, et al. Frequently deleted loci on chromosome 9 may harbor several tumor suppressor genes in human renal cell carcinoma. J Urol 2001;166:1088–1092.
Presti Jr JC, Wilhelm M, Reuter V, et al. Allelic loss on chromosomes 8 and 9 correlates with clinical outcome in locally advanced clear cell carcinoma of the kidney. J Urol 2002;167:1464–1468.
Kinoshita H, Yamada H, Ogawa O, et al. Contribution of chromosome 9p21–22 deletion to the progression of human renal cell carcinoma. Jpn J Cancer Res 1995;86:795–799.
Gunawan B, Huber W, Holtrup M, et al. Prognostic impacts of cytogenetic findings in clear cell renal cell carcinoma: gain of 5q31-qter predicts a distinct clinical phenotype with favorable prognosis. Cancer Res 2001;61:7731–7738.
Acknowledgements
We acknowledge Fondazione Cassa di Risparmio di Verona; Diagnostica molecolare in oncologia; Ministero Istruzione, Università e Ricerca (MIUR); Banco di Sardegna. This work has been accepted in part for presentation at the 96th Annual Meeting of the US and Canadian Academy of Pathology, San Diego, CA, USA, 24–30 March 2007.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brunelli, M., Eccher, A., Gobbo, S. et al. Loss of chromosome 9p is an independent prognostic factor in patients with clear cell renal cell carcinoma. Mod Pathol 21, 1–6 (2008). https://doi.org/10.1038/modpathol.3800967
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800967
Keywords
This article is cited by
-
Characterisation of tumor microenvironment and prevalence of CD274/PD-L1 genetic alterations difference in colorectal Cancer
BMC Cancer (2023)
-
A Meta-Analysis Evaluating Clinical Outcomes of Patients with Renal Cell Carcinoma Harboring Chromosome 9P Loss
Molecular Diagnosis & Therapy (2019)
-
Chromosome 9p deletion in clear cell renal cell carcinoma predicts recurrence and survival following surgery
British Journal of Cancer (2014)
-
Renal cell carcinoma with smooth muscle stroma lacks chromosome 3p and VHL alterations
Modern Pathology (2014)
-
Renal Cell Carcinoma Deep Sequencing: Recent Developments
Current Oncology Reports (2012)