Abstract
PCA3 is a specific marker of prostatic carcinoma. However, PCA3 has been detected only at RNA level and a corresponding PCA3 protein has never been identified. The aim of this study was to develop a technique capable of detecting PCA3 RNA on histology sections and to assess the cellular location of the molecule. Forty-eight formalin-fixed paraffin-embedded blocks of prostatectomy specimens were selected for PCA3 detection by in situ hybridization by both radioactive and chromogenic methods. Of the 48 sections, 28 contained prostatic adenocarcinoma and 20 had benign tissue located distant from the tumor. Using the radioactive detection method, 26 of 28 available cases (93%) of cancers presented at least focal cytoplasmic PCA3 expression. The benign glands located in proximity of the cancer presented PCA3 expression in eight (29%) cases, whereas those situated distant to the tumor showed focal expression in 2 of 20 (10%) cases only. High-grade prostatic intraepithelial neoplasia (HGPIN) expressed PCA3 in 25 of 26 (96%) cases. With the chromogenic detection method, 22 of the 24 interpretable cases (92%) of cancers had at least focal cytoplasmic staining. Benign glands located close to neoplastic glands expressed PCA3 in 8 (33%) cases, but none of those distant to the tumor expressed the marker. HGPIN was positive in 17 of 24 (71%) cases. The sensitivity, specificity, positive predictive value and negative predictive value for the detection of cancer were 93, 79, 71 and 95% for the radioactive detective method and 92, 80, 71 and 95% for the chromogenic detection method, respectively. Our study shows that PCA3 RNA is expressed by most prostate cancers and HGPIN. Normal glands rarely express the marker, except those located in immediate proximity of neoplastic glands, suggesting the presence of precursor molecular changes.
Similar content being viewed by others
Main
Prostate cancer is the most frequent malignancy in men of all races and the second leading cause of death from cancer in North America, surpassed only by lung cancer.
Due to increased accuracy of diagnostic techniques and extended screening programs, the neoplastic disease, in general, tends to be discovered at an early and potentially curable stage. The overall mortality from prostate cancer declined for the first time in the mid-1990s, after more than 60 years of an increasing trend.1, 2 This is largely attributable to a declining detection of late-stage disease and increased locally and regionally limited disease, likely due to the extensive screening programs in the prostate-specific antigen (PSA) era. Concurrently, this decline is associated with a reduced percentage of patients treated with conservative measures as first-line therapy.
Sextant prostate biopsy yields 15–34% negative results3 in patients later proven to have cancer on subsequent biopsies. The optimal protocol regarding the number of cores and their localization is still a matter of debate.4 When total serum PSA level ranges between 4 and 10 ng/ml, transrectal ultrasound-guided biopsy detects only 22% of cancer at the first biopsy, and repeated second, third and fourth biopsies add 10, 5 and 4% of positive cases, respectively.5 This is explained, in part, by the lack of detection of cancer on biopsies due to the small number of atypical glands. Another reason is the lack of criteria to detect abnormal glands that have part but not all the criteria of malignancy. Therefore, there is a need to identify new markers specific for prostate neoplasia that may help to detect early-stage cancers and to discriminate malignant from benign tissue on difficult-to-interpret biopsies.
PCA3 (also known as DD3 or PCA3DD3) is one of the most prostate cancer-specific genes ever described. It is overexpressed in the majority of prostate cancers including distant metastases, expressed at a negligible level in nonmalignant prostate tissue and not expressed in other normal or malignant tissues.6, 7, 8
As PCA3 is a noncoding gene, there is no corresponding protein eventually detectable by immunohistochemistry.9 Also, the cellular location of the molecule has never been identified. In this study, we present the development of a technique capable of detecting and assessing the cellular location of PCA3 RNA in prostate paraffin-embedded tissue by in situ hybridization (ISH).
Materials and methods
The study population consisted of 48 patients with prostate adenocarcinoma submitted to radical prostatectomy at L’Hôtel-Dieu de Québec, Quebec, Canada, between 1999 and 2003. Written informed consent was obtained from all study participants. The protocol was approved by the Institutional Ethical Research Committee.
Of those 48 cases, we selected 28 paraffin-embedded blocks containing a mixture of cancer and normal glands. The benign glands present on these sections were considered as being located close to the tumor. In 20 separate blocks, only normal tissue distant from the tumor was present. The presence of high-grade prostate intraepithelial neoplasia (HGPIN) was assessed on all sections.
ISH with Radioactive Detection Probes
ISH was performed using a 460-base, 35S-UTP-labeled probe, generated by a T7 transcription from a PCA3 construct subcloned in pSPT-19 plasmid (Roche Diagnostics Corp., Indianapolis, IN, USA). The probe included the 3′ portions of exons 1 and 3 and the 5′ portion of exon 4 of the PCA3 RNA (Figure 1). Briefly, 4 μm sections were cut from the paraffin-embedded blocks in an RNase-free environment, deparaffinized, rehydrated, washed and then digested in 2 μg/ml Proteinase K solution (Roche) at 37°C for 30 min. After washing, the slides were treated in an acetylation bath for 10 min at room temperature, then rewashed and dried. The slides were subsequently hybridized overnight at 58°C with 50 μl of a 4 × 104 CPM/μl labeled probe. The next day, after a stringent post-hybridization washing, the sections were exposed to an autoradiography emulsion and stored in the dark at 4°C for 2 weeks until developed. Finally, slides were counterstained with toluidine blue and coverslips were placed on the slides.
Schematic representation of the PCA3 transcription unit. The PCA3 gene consists of four exons where exon 2 is often skipped by alternative splicing. Three alternative polyadenylations can occur in exon 4 (4a, 4b and 4c). The 460 bp ISH probe shown here targets exons 1, 3 and 4 of the PCA3 RNA and corresponds to positions 95–120 and 286–719 in GenBank accession number AF103907.
ISH with Chromogenic Detection Probes
ISH was performed using a 460-base digoxigenin-labeled probe generated by PCR from the plasmid used in the radioactive technique. Four-micrometer-thick tissue sections were deparaffinized, rehydrated, pretreated in 0.2 M hydrochloric acid and then digested with a 20 μg/ml Proteinase K solution at 37°C for 10 min. After washing, the slides were fixed in neutral buffered formalin for 10 min, treated for 10 min with 0.2% glycine, washed again and immersed in an acetylation bath for 10 min at room temperature, and then rewashed and dried. The slides were subsequently incubated for 1 h at 43°C with the hybridization buffer and hybridized overnight at 37°C with the 2 μg/ml digoxigenin-labeled probe. The next day, after stringent post-hybridization washing, digoxigenin-labeled probes were detected using a mouse monoclonal anti-digoxigenin antibody (Roche Diagnostics), at 1:500 dilution and 1 h incubation time. Subsequently, to amplify and visualize the positive signal, we used the Catalyzed Signal Amplification System (Dako, CA, USA; CSA code K1500) following the manufacturer's instructions and using 3,3′ diaminobenzidine as the chromogen. Finally, the slides were counterstained with hematoxylin, dehydrated and coverslips were placed on the slides.
The positive control included in each run consisted of slides where no stringent post-hybridization washing was performed. The negative control consisted of slides where the radiolabeled probe or the primary anti-digoxigenin antibody was omitted.
Interpretation and Statistical Analyses
The percentage of cells expressing the marker was evaluated for each case and with each method. The presence of a single positive cell was defined as positive. Negative was defined as the complete absence of expression. The specificity, sensitivity, positive predictive value and negative predictive value were calculated for each detection method. The Pearson's correlation along with P-value was calculated to search for a linear relationship between percentage of positive cells and age, pre-surgery PSA, Gleason grade and stage (the stage was converted to a continuous variable). A P-value of less than 0.05 was considered significant.
Results
Clinical characteristics for the study population are summarized in Table 1. The median patient age was 65 years. The median pre-surgery PSA value was 9.7. All patients had a conventional prostate adenocarcinoma. Twelve (43%) patients had a stage pT3 tumor and four (14%) had lymph node metastases. No patient presented distant metastases.
ISH by Radioactive Method
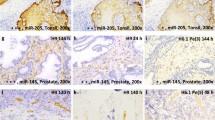
Marker expression for each case is depicted in Table 1. Of the 28 tumor specimens, PCA3 RNA expression was positive in 26 (93%) cases and 2 (7%) were negative. In all positive cases, the PCA3 expression was located in the cytoplasm of cancer cells (Figure 2a and b). The number of positive cells ranged from 10 to 100%, and 22 of 28 available cases (79%) had more than 10% of cells expressing the marker (Table 1). There was no association between the presence or absence of expression or the extent of PCA3 RNA expression and the age, stage, the Gleason score or pre-surgery PSA (Table 2). The benign glands located close to neoplastic glands exhibited focal cytoplasmic expression in eight (29%) cases. Overall, the radioactive method presented a sensitivity at detecting tumors of 93%, a specificity of 79%, a positive predictive value of 71% and a negative predictive value of 95%. Of the 20 specimen blocks with normal tissue distant from the tumor, focal PCA3 expression was present in 2 (10%) cases only. HGPIN was present in 26 of the 48 examined sections. PCA3 expression was present in 25 (96%) of 26 cases (Figure 2c). One case (4%) was negative. The stromal cells were negative in all cases.
ISH by Chromogenic Method
Adequate and interpretable tissue was available for 24 out of 28 tumor blocks used for radioactive detection. Detailed data are presented in Table 1. Two cases (8%) had PCA3-negative expression. The other 22 (92%) cases had PCA3 expression detected in the cytoplasm (Figure 3a and b). The number of positive cells ranged from 5 to 100%, and 18 of 24 available cases (75%) had more than 10% of cells expressing the marker (Table 1). The two tumors found negative by the radioactive technique were also negative by the chromogenic method. There was no association between the presence of PCA3 RNA or the level of expression and the age, stage, the Gleason score or pre-surgery PSA (Table 2). Normal glands from the tumor blocks presented focal PCA3 expression in eight (33%) cases. All 20 normal tissues remote from the tumor were negative for the marker. HGPIN was present in 24 cases. The marker was expressed (Figure 3c) in 17 (71%) cases. Stromal cells were negative in all cases.
Globally, the chromogenic method of PCA3 detection in cancer cells presented a sensitivity of 92%, a specificity of 80%, a positive predictive value of 71% and a negative predictive value of 95%.
Discussion
PCA3 was described for the first time by Bussemakers et al7 in 1999 as a highly specific marker of prostate carcinoma. One year later, the promoter was sequenced by Verhaegh et al.6 According to Gandini et al,10 only the splicing variant of exon 4 is specific for prostate cancer. As PCA3 is highly expressed in prostate cancer compared to normal prostate and negative in other normal and malignant tissues, this molecule is regarded as a potential target for therapy11, 12 or an adjunct to diagnosis.13
Using formalin-fixed paraffin-embedded prostate adenocarcinoma, Balcerczak et al14 found by reverse transcriptase-polymerase chain reaction (RT-PCR) a high expression of PCA3 mRNA in well- and moderately differentiated tumors and a low expression in high-grade tumors. However, Bialkowska-Hobrzanska et al,15 using the same technique on paraffin-embedded tissue, found that PCA3 is highly expressed in prostate cancers, irrespective of stage or Gleason score.
Landers et al16 confirmed that PCA3 is overexpressed up to 140-fold in cancer samples compared to benign prostate hyperplasia and that a combination of PCA3, GalNAc-T3, hepsin and PSMA can distinguish cancer tissue from benign prostatic hyperplasia (BPH) with a 100% accuracy rate. In a study on nine prostate-related transcripts, PCA3 was found as the best single marker for the detection of prostate carcinoma.17
Using laser microdissection, Thelen et al18 detected PCA3 RNA by RT-PCR in all analyzed prostate cancer samples. Petrovics et al19 showed that, using this technology, a combination of PCA3, α-methyl acyl CoA racemase (AMACR) and ETS-related gene is capable of detecting virtually all prostate cancers. The regulatory mechanism of PCA3 secretion is unknown. However, von Bokhoven et al20 and Schalken et al9 suggest that PCA3 expression could be regulated via the androgen receptor pathways.
A number of clinical translational studies investigating the role of PCA3 as a marker of prostate cancer have been published. Jung et al21 detected PCA3 mRNA in blood by RT-PCR. By a similar technique, Marangoni et al22 have used PCA3 as a marker to detect circulating prostatic cancer cells in blood.
On a large cohort of 517 patients, Fradet et al23 demonstrated that uPM3, a test detecting PCA3 and PSA mRNA in urine samples, can be a useful adjunct to current methods for the detection of early prostate cancer. The overall sensitivity and specificity were 66 and 89%, respectively; the positive predictive value was 75% and the negative predictive value was 84%. In similar studies, Tinzl et al24 tested urine samples with the uPM3 test on a cohort of 201 patients. The sensitivity, specificity, positive predictive value and negative predictive value were 82, 76, 67 and 87%, respectively. Hessels et al25 investigated PCA3 expression by quantitative RT-PCR assay in urine samples collected after prostatic massage. The test sensitivity, specificity and negative predictive value were 67, 83 and 90%, respectively. In a recent study by Bostwick et al,26 PCA3 RNA was detected by the uPM3 test in urine samples of patients with prostate cancer irrespective of tumor volume, location, extent and grade. A newly developed research-use only urine test detecting PCA3 RNA in urine (Aptima PCA3, Gen-Probe Inc., San Diego, CA, USA) showed a global sensitivity and specificity of 69 and 79%, respectively.27
However, no study published to date has attempted to localize the marker in tissue sections. Our study showed for the first time a sensitivity of 92–93% for PCA3 to detect prostate cancer on tissue sections, using either a radioactive or a nonradioactive method. The advantage of the nonradioactive approach is its possible use in a routine surgical pathology laboratory, whereas the radioactive method requires special precautions and equipment. Two tumors were negative by both the radioactive and the chromogenic techniques. This suggests that technical difficulties such as long fixation time or tissue manipulation in non-RNase-free environment may be responsible for this lack of marker expression rather than a true negativity.
We demonstrated for the first time that PCA3 is located in the cytoplasm of cancer cells and is absent in stromal cells. Furthermore, we also demonstrated that PCA3 is significantly expressed in HGPIN. This expression is not surprising, considering that HGPIN is regarded as a precursor of prostate cancer. Finally, some normal-appearing glands situated in the immediate vicinity of cancer glands express PCA3, which is virtually absent in remotely located normal glands. This suggests a possible carcinogenic field effect in normal glands situated close to the tumor. However, although this marker has the potential to become an adjunct in the diagnosis of prostatic carcinoma, further studies should assess PCA3 RNA expression in benign histopathologic entities and prostatic cancer mimics, including atrophy, inflamed benign glands, glandular hyperplasia in BPH nodules, basal cell hyperplasia, seminal vesicle glands and atypical adenomatous hyperplasia. Analyses should also determine whether PCA3 provides information beyond that of currently used basal cell markers and AMACR.
Our findings on the cellular location of PCA3 differ from those published by Schalken et al.9 Although the authors acknowledge that the exact location of the transcripts has not yet been clearly established, they mention that they found indications that most PCA3 transcripts are located in the nucleus. The method used to reach this conclusion is, however, not mentioned. In our study, PCA3 was unequivocally expressed in cell cytoplasm and was not detected in the nucleus using either technique.
In conclusion, we present the development of radioactive and chromogenic ISH techniques capable of detecting and localizing PCA3 RNA at the cellular level. We found that the marker is expressed in the cytoplasm of cancer cells, HGPIN and some normal cells and absent in stromal cells.
Accession codes
References
Edwards BK, Brown ML, Wingo PA, et al. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst 2005;97:1407–1427.
Wingo PA, Cardinez CJ, Landis SH, et al. Long-term trends in cancer mortality in the United States, 1930–1998. Cancer 2003;97:3133–3275.
Terris MK . Sensitivity and specificity of sextant biopsies in the detection of prostate cancer: preliminary report. Urology 1999;54:486–489.
Naughton CK, Smith DS, Humphrey PA, et al. Clinical and pathologic tumor characteristics of prostate cancer as a function of the number of biopsy cores: a retrospective study. Urology 1998;52:808–813.
Djavan B, Milani S, Remzi M . Prostate biopsy: who, how and when. An update. Can J Urol 2005;12(Suppl 1):44–48; discussion 99–100.
Verhaegh GW, van Bokhoven A, Smit F, et al. Isolation and characterization of the promoter of the human prostate cancer-specific DD3 gene. J Biol Chem 2000;275:37496–37503.
Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999;59:5975–5979.
Schalken JA . Validation of molecular targets in prostate cancer. BJU Int 2005;96:23–29.
Schalken JA, Hessels D, Verhaegh G . New targets for therapy in prostate cancer: differential display code 3 (DD3(PCA3)), a highly prostate cancer-specific gene. Urology 2003;62:34–43.
Gandini O, Santulli M, Cardillo MR, et al. Correspondence re: JB de Kok et al., DD3, a very sensitive and specific marker to detect prostate tumors. Cancer Res 2002;62:2695–2698. Cancer Res 2003; 63: 4747–4749.
Bussemakers MJ . Changes in gene expression and targets for therapy. Eur Urol 1999;35:408–412.
Schalken J . Molecular diagnostics and therapy of prostate cancer: new avenues. Eur Urol 1998;34(Suppl 3):3–6.
de Kok JB, Verhaegh GW, Roelofs RW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res 2002;62:2695–2698.
Balcerczak E, Mirowski M, Sasor A, et al. Expression of p65, DD3 and c-erbB2 genes in prostate cancer. Neoplasma 2003;50:97–101.
Bialkowska-Hobrzanska H, Driman DK, Fletcher R, et al. Expression of human telomerase reverse transcriptase, Survivin, DD3 and PCGEM1 messenger RNA in archival prostate carcinoma tissue. Can J Urol 2006;13:2967–2974.
Landers KA, Burger MJ, Tebay MA, et al. Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int J Cancer 2005;114:950–956.
Schmidt U, Fuessel S, Koch R, et al. Quantitative multi-gene expression profiling of primary prostate cancer. Prostate 2006;66:1521–1534.
Thelen P, Burfeind P, Grzmil M, et al. cDNA microarray analysis with amplified RNA after isolation of intact cellular RNA from neoplastic and non-neoplastic prostate tissue separated by laser microdissections. Int J Oncol 2004;24:1085–1092.
Petrovics G, Liu A, Shaheduzzaman S, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 2005;24:3847–3852.
van Bokhoven A, Varella-Garcia M, Korch C, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate 2003;57:205–225.
Jung M, Xu C, Spethmann J, et al. Re: Hessels D, Klein Gunnewiek JMT, van Oort I, Karthaus HFM, van Leenders GJL, van Balken B, Kiemeney LA, Witjes JA, Schalken JA. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 2003;44:8–16. Eur Urol 2004; 46: 271–272.
Marangoni K, Neves AF, Cardoso AM, et al. The endothelial nitric oxide synthase Glu-298-Asp polymorphism and its mRNA expression in the peripheral blood of patients with prostate cancer and benign prostatic hyperplasia. Cancer Detect Prev 2006;30:7–13.
Fradet Y, Saad F, Aprikian A, et al. uPM3, a new molecular urine test for the detection of prostate cancer. Urology 2004;64:311–315.
Tinzl M, Marberger M, Horvath S, et al. DD3PCA3 RNA analysis in urine—a new perspective for detecting prostate cancer. Eur Urol 2004;46:182–186.
Hessels D, Klein Gunnewiek JM, van OI, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 2003;44:8–15.
Bostwick DG, Gould VE, Qian J, et al. Prostate cancer detected by uPM3: radical prostatectomy findings. Mod Pathol 2006;19:630–633.
Groskopf J, Aubin SM, Deras IL, et al. Aptima PCA3 Molecular Urine Test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem 2006;52:1089–1095.
Acknowledgements
We thank Timothy Holzer, Martin Beaulieu and Lyson Piché for their help in reviewing the manuscript and Francois Harel for the statistical calculation. This study was sponsored by Diagnocure Inc., QC, Canada. The results were partially presented as a poster at the United States & Canadian Academy of Pathology (USCAP) Annual Meeting in February 2005 in San Antonio, TX, USA.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The initial discovery of PCA3 was the result of a joint effort between the Radboud University Medical Centre in Nijmegen, The Netherlands, and the John Hopkins University in Baltimore, Maryland, USA. DiagnoCure owns worldwide exclusive rights to the diagnostic, prognostic and therapeutic applications of PCA3, which are protected through patents and patent applications worldwide. In 2003, DiagnoCure has licensed the diagnostic applications to Gen-Probe. Dr Fradet is also Vice-President of DiagnoCure Inc. and Guillaume Beaudry is an employee of the company. There is no duality of interest for other authors.
Rights and permissions
About this article
Cite this article
Popa, I., Fradet, Y., Beaudry, G. et al. Identification of PCA3 (DD3) in prostatic carcinoma by in situ hybridization. Mod Pathol 20, 1121–1127 (2007). https://doi.org/10.1038/modpathol.3800963
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800963
Keywords
This article is cited by
-
Long non-coding RNAs involved in autophagy regulation
Cell Death & Disease (2017)
-
Global expression of AMACR transcripts predicts risk for prostate cancer – a systematic comparison of AMACR protein and mRNA expression in cancerous and noncancerous prostate
BMC Urology (2016)
-
Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cancer cells
Tumor Biology (2016)
-
Altered PCA3 and TMPRSS2-ERG expression in histologically benign regions of cancerous prostates: a systematic, quantitative mRNA analysis in five prostates
BMC Urology (2015)
-
Prostate-specific RNA aptamer: promising nucleic acid antibody-like cancer detection
Scientific Reports (2015)