Abstract
Human homolog of murine double minute 2 (HDM2) and HDM4 (or HDMX) are negative regulators of p53. HDM4 has not been assessed in precursor B (pre-B) lymphoblastic leukemia (ALL). We examined bone marrow samples obtained at time of diagnosis from 55 adults with pre-B ALL. A tissue microarray composed of 2 cores per specimen was constructed and immunohistochemical techniques were used to assess HDM4, HDM2, p53, and p21. HDM4 was expressed in 39 of 49 (80%) cases. HDM2 was expressed in 14 of 54 (26%). All HDM2-positive cases were also positive for HDM4 (P<0.05). We confirmed expression of HDM4 and HDM4 variants by Western blotting and sequencing of reverse transcription-polymerase chain reaction products in a subset of ALL tumors. Results were correlated with the presence of the Philadelphia chromosome (Ph). p53 (P<0.05) and p21 (P<0.001) were expressed significantly more often in Ph+ pre-B ALL. HDM4 and HDM2 showed no correlation with Ph status. HDM4 expression in most cases of adult pre-B ALL suggests that HDM4 is a potential therapeutic target.
Similar content being viewed by others
Main
There has been little progress in prolonging the survival of adult patients with precursor B (pre-B) lymphoblastic leukemia (ALL) using empiric chemotherapy regimens. The long-term survival rate in these patients is only 30%.1, 2, 3 An improved understanding of molecular events in this disease is clearly needed.
The human homolog of murine double minute 2, (HDM2) functions as a negative regulator of p53 through inhibition of transcriptional activation and degradation via its E3 ubiquitin ligase activity.4 HDM4 was initially identified in mice as Mdm4, shown to be a p53-binding protein, and later as a Mdm2 partner.5, 6 HDM4 and HDM2 mRNA encode structurally related proteins of 490 and 491 amino acids, respectively. Deletion of either Mdm2 or Mdm4 in mice results in embryonal lethality, which can be reversed by deletion of p53.7, 8, 9 These findings strongly indicate that Mdm2 and Mdm4 have important roles in the regulation of p53 activity in mice in vivo. Recent data have also shown that HDM4 stabilizes the p53-HDM2 complex and increases the function of the E3 ubiquitin ligase of HDM2, resulting in degradation of p53 through a proteasome-dependent pathway.10, 11 HDM2 also shuttles HDM4 into the nucleus, as HDM4 normally exists only within the cytoplasm because it lacks a nuclear localization signal sequence.12 In mice, overexpression of Mdm4 allows immortalization and neoplastic transformation of primary mouse embryonic fibroblasts.13 The importance of Mdm4 in negative regulation of p53 suggests that HDM4 may be a new drug target for therapy in cancer patients.
The p53 gene has a myriad of functions and activates a number of genes involved in cell cycle arrest and apoptosis. There is evidence in the literature to suggest that p53 deregulation or inactivation occurs in a subset of pre-B ALL cases. In one study, the p53 gene was inactivated by mutation, deletion, or promoter methylation in 8.8, 7.5, and 32% of ALL cases, respectively.14, 15 Another study showed that p53 gene mutations are relatively uncommon in pre-B ALL at initial diagnosis, 3%, but more common at time of relapse, 30%.16 Alternatively, it has been suggested that inactivation of wild-type p53 protein frequently occurs through binding to its negative regulators, HDM2 and HDM4.
To gain insight into the role of HDM4 in the pathogenesis of adult pre-B ALL, we immunohistochemically assessed for expression of HDM4, HDM2, p53, and p21 in a group of patients treated in a standardized fashion.3, 17 Our results show that HDM4 is usually expressed, and the p53 pathway is usually altered in adult pre-B ALL.
Materials and methods
We studied 55 adult patients with de novo pre-B ALL treated at The University of Texas, MD Anderson Cancer Center (MDACC). All subjects gave their written informed consent, and the Institutional Review Board approved the protocol. The diagnosis of pre-B ALL was based on a combination of clinical, morphologic, laboratory, and immunophenotypic criteria as defined previously.17 Each patient was treated with the HCVAD regimen (hyperfractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone).
For construction of the tissue microarray, hematoxylin and eosin-stained slides prepared from paraffin-embedded, formalin-fixed blocks of bone marrow biopsy specimens were used to define diagnostic areas. Two representative 0.6-mm cores were then obtained from each sample and inserted in a grid pattern into a recipient paraffin block by using a tissue arrayer (Beecher Instruments, Silver Spring, MD, USA) as described elsewhere.18 We stained sections from each tissue microarray using immunohistochemical methods described previously.18 The antibodies used were specific for p53 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA), p21 (1:75; NeoMarkers, Fremont, CA, USA), HDM2 (1:75; Santa Cruz Biotechnology), and HDM4 (1:200; rabbit polyclonal against human recombinant glutathione-S-transferase (GST)-MDMXΔ109-198 protein antibody generated by Dr Guillermina Lozano, manuscript submitted).
Cases were considered positive if 10% or more of the tumor cells were stained. Images of the tissue microarray were acquired and stored in an on-line image archive linked to the array database as previously described.18
Western Blot Analysis of HDM4 in Pre-B ALL Specimens
To test whether HDM4 is expressed, cell extracts were prepared19 from peripheral blood specimens obtained from adults with pre-B ALL who had high white blood cell counts and greater than 90% blasts. The proteins were resolved on 10% sodium dodecyl sulfate-polyacrylamide gels. After electrophoresis, proteins were electrotransferred to nitrocellulose filters (Hybond-ECL; Amersham Corporation, Arlington Heights, IL, USA), probed with antibody specific for HDM4 (anti-GST-MDMXΔ109-198 recombinant protein, 1:1,000, rabbit polyclonal), and detected by chemiluminescence (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK). After initial probing, blots were reprobed with an antibody specific for actin (1:1000, mouse monoclonal, Sigma, St Louis, MD, USA) to assess protein loading.
Reverse Transcriptase-Polymerase Chain Reaction Analysis and Direct Sequencing of PCR Products
To characterize the variants of HDM4, reverse transcriptase-polymerase chain reaction (RT-PCR) assays were performed as described elsewhere.19 Peripheral blood specimens obtained from adults with pre-B ALL as described above were used for analysis of HDM4. Briefly, total RNA was extracted from 2 ml of EDTA-anticoagulated blood specimens using the Trizol method (Life Technologies, Rockville, MD, USA). After treatment with RNAse-free DNAse, total RNA was used for complementary DNA synthesis using an oligo-dT primer. Appropriate sets of primers were modified and designed for PCR amplification according to the sequences reported in the literature.19 Amplified products were run on an 8% polyacrylamide gel and detected with ethidium bromide stain. The amplified bands were then submitted for direct sequence analysis. Sequencing was performed using ABI Big Dye terminator cycle sequencing chemistry (Applied Biosystems, Foster City, CA, USA), at the DNA Analysis Core Facility of MDACC.
P53 Gene Mutation Analysis
Purified genomic DNA specimens were available from five of 15 pre-B ALL specimens that intensely expressed p53. Genomic DNA was extracted from 2 ml of EDTA anticoagulated bone marrow aspirate material using standard DNA extraction methods. These specimens were then analyzed for p53 mutation using the commercially available VariantSEQr kit (Applied Biosystems) at the DNA Analysis Core Facility of MDACC. Briefly, 12.5 ng of genomic DNA is used for each PCR. A total of 17 pairs of primers were used for amplification of both exons and introns of p53 genomic DNA, and then fluorescently labeled amplified products were analyzed using universal sequencing primers, BigDye Terminator v3.0 chemistry, and the Applied Biosystems 3730 Genetic Analyzer. These analyses cover more than 99% of the p53 gene mutations reported in the literature.
Statistics
Fisher's exact test was used to investigate the association between expression of HDM4 and that of HDM2, p53, and p21, as well as the association between Ph status and HDM4, HDM2, p53, and p21 expression. Also, the associations of overall survival, Ph status, and expression of HDM4, HDM2, p53, and p21 were explored. Kaplan–Meier survival curves and the log-rank test were used to determine differences in survival distribution by Ph status and the biomarkers of interest. Tests for the association between biomarkers and survival were stratified by Ph status because it is highly associated with survival. The Cox proportional hazards model was used to relate overall survival with the interaction between Ph status and the biomarkers of interest. All grouped P-values were two-sided, and considered to be statistically significant if P≤0.05.
Results
Clinicopathologic Characteristics
Clinicopathologic and cytogenetic data for the 55 study patients are summarized in Table 1. The follow-up duration ranged from 2 to 675 weeks. The estimated median overall survival duration was 75 weeks (95% confidence interval (CI), 62–138 weeks). Fifty-one patients had a response to therapy, and 41 patients eventually relapsed. The median disease-free survival duration was 59 weeks (95% CI, 43–114 weeks).
Frequency and Association of HDM4, HDM2, p53 and p21 Expression in Adult Pre-B ALL
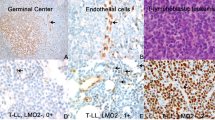
HDM4 was expressed in 39 of 49 (80%) cases of pre-B ALL, p53 was expressed in 20 of 53 (38%), p21 was expressed in 17 of 51 (33%), and HDM2 was expressed in 14 of 54 (26%). Most of HDM4+ cases showed strong nuclear staining for HDM4 in more than 75% of leukemia blasts, even in cases that were negative for HDM2 staining. Representative sections of the tissue microarray with immunostaining results are shown in Figure 1. Table 2 summarizes the associations between HDM4 and p21, p53, and HDM2. All 14 cases of pre-B ALL positive for HDM2 were also positive for HDM4 and this association was significant (P<0.05) (Table 3). We detected no statistically significant associations between expression of HDM4 and p53 or p21.
Association between Protein Expression and Ph Status
p53 was expressed more often in Ph+ cases (10/17 Ph+ vs 10/36 Ph−; P<0.05), as was p21 (11/17 Ph+ vs 6/34 Ph−; P<0.001). HDM4 and HDM2 expression did not correlate with Ph status (Table 2).
Association between Protein Expression and Cytogenetic Abnormalities
Five cases had abnormalities of chromosome 1, but none involved 1q32 (the gene locus for HDM4). Four of these cases were strongly and uniformly positive for HDM4, whereas one was negative. One case had a chromosomal translocation involving 12q12 (near the gene locus for HDM2). This case was immunohistochemically negative for HDM2. One case with addition of 17p13 (the gene locus for p53) showed high-intensity staining for p53.
Impact of Ph Status, HDM4, HDM2, p53, and p21 Expression on Survival
The median survival duration for patients with Ph− and Ph+ pre-B ALL was 138 weeks and 48 weeks, respectively (P<0.001) (Table 1, Figure 2). No single immunohistochemical marker demonstrated an impact on survival (Table 4, Figures 2 and 3). Figure 2 displays the overall survival duration and associated 95% CIs according to Ph, p21, p53, HDM2, and HDM4. Forty-two patients died during follow-up. In the HDM2-negative group, expression of HDM4 did not show a significant effect on survival duration. The Cox proportional hazards model, used to test the interaction between Ph status and the biomarkers on survival, detected no significant interactions.
Analysis of the Control Group
We used six cases of precursor T-cell ALL as a control group. All six patients were Ph−. One case was positive for p53, none were positive for HDM2, five were positive for HDM4, and one was positive for p21. We also analyzed normal bone marrow; erythroid precursors were positive for HDM4 with a nuclear pattern of staining. All normal bone marrow elements were negative for p53, p21, and HDM2.
Western Blot and RT-PCR Analyses of HDM4
Two specimens from pre-B ALL patients showed the presence of a normal sized HDM4 protein of 80 kDa, while one specimen demonstrated two bands, with the second band ∼30 kDa. Two control cell lines, Raji (human Burkitt lymphoma) and Molt (human precursor T-cell ALL), had only the small ∼30 kDa bands (Figure 4a). The results of RT-PCR from two pre-B ALL specimens confirmed the presence of two splicing forms of HDM4 mRNA (Figure 4b). The RT-PCR products were sequenced and the presence of a HDM4 variant with a short internal deletion of 68 bp was confirmed (Figure 4c).
Variants of HDM4 in pre-B ALL. (a) Western blot analysis showed HDM4 proteins in human pre-T ALL (Molt) and human Burkitt lymphoma (Raji) cell lines and pre-B ALL specimens. (b) The presence of different splicing forms, HDM4 and HDM4-S, were confirmed by RT-PCR analysis. (c) Direct sequencing of the RT-PCR products confirmed the presence of both the HDM4 and HDM4-S forms. The underlined 68 nucleotides (upper) are the sequence that will be spliced out in the HDM4-S (lower). The arrowed lines point at the new junction in the HDM4-S.
P53 Gene Mutation Analysis in Pre-B ALL Specimens
One of the five analyzed cases showed a missense mutation in exon 7 and a silent mutation in exon 4 of the p53 gene. Three cases each had two silent mutations in exons 4, and 11 or intron 9 (Table 5). One case had a single silent mutation in exon 4.
Discussion
In this study, we assessed expression of HDM4, HDM2, p53, and p21 in adult pre-B ALL. Whereas p21, p53, and HDM2 were evaluated individually in previous studies, this is the first study to analyze HDM4 expression in adult pre-B ALL. The goals of this study were two. First, we wanted to assess the frequency of HDM4 expression in adult pre-B ALL. Second, we wanted to assess how expression of these proteins may be inter-related.
HDM4 is frequently expressed in adult pre-B ALL, in 80% of cases in this study. Even though HDM4 and HDM2 are homologs and both inactivate p53, there was a significant correlation between HMD4 and HMD2 expression. None of the cases negative for HDM4 were positive for HDM2. Thus, these proteins may act cooperatively to inhibit p53 activity. Our data suggest that HDM4 is a potential therapeutic target and support the hypothesis that HDM4 stabilizes the p53-HDM2 complex in a subset of pre-B ALL.
HDM4 occupies a unique and very critical position in the regulation of the p53. HDM4 is the only inhibitor of p53 not also regulated by p53 itself. Several splicing variants of HDM4, including HDM4-A, HDM4-G, HDM4-S (or HDM4-E), a human homolog of MDMX-S in the mouse, have been described.20, 21 Because of a lack of exon 9 sequences encoding the acidic domain, HDM4-A cannot stabilize HDM2 and is particularly susceptible to HDM2-triggered degradation. Also, HDM4-G lacks exons 3–6 and cannot bind to p53. Meanwhile, HDM4-S is a truncated HDM4 protein and represents only the p53-binding domain. It is generated by a short internal deletion of 68 bp that produces a shift in the reading frame after codon 114, and an early stop codon at amino-acid residue 127. HDM4-S binds tightly to p53 and inhibits p53-mediated transcriptional activation that induces apoptosis. In addition, another spliced form of HDM4, named HDMX211, was also recently isolated from the thyroid tumor cell line, ARO. This spliced form contains exons 1 and 2, and most of exon 11. HDMX211 binds and stabilizes HDM2 and also counteracts HDM2-mediated p53 degradation.22
Interestingly, Western blot analysis of one of three ALL cases showed the presence of an additional band at ∼30 kDa, suggesting that some pre-B ALL cases have both HDM4 and HDM4-S forms. The antibody against HDM4 and primers for RT-PCR used in this study will not detect the protein or mRNA of the HDMX211 variant according to the reported sequence. Recently, Bartel et al23 reported that overexpression of the HDM4-S transcript and HDM4 gene amplification are important prognostic markers for soft tissue sarcoma. Further investigation is needed to clarify the importance of HDM4-S and HDMX211 in pre-B ALL.
HDM4 splicing variants as well as full-length HDM4 are potential therapeutic targets for small molecule inhibitors that can interfere with interactions among HDM4, HDM2, and p53. Nutlin-3, an antagonist of p53-HDM2 interaction, recently has been shown to synergistically improve response of tumor cells to chemotherapy.24, 25 Although the p53 binding domains of HDM2 and HDM4 are well preserved, Nutlin-3 does not effectively disrupt the interaction between p53 and HDM4.26 The status of HDM4 dictates the response of tumor cells to Nutlin-3, indicating the HDM4 most likely plays a pivotal role in any therapeutic strategy targeting p53 reactivation in some cases.27 Ramos et al28 and Riemenschneider et al29 have shown that aberrant expression of HDM4 in tumor cell lines correlates with expression of wild-type p53, and that interference between HDM2 and p53 causes cell cycle arrest and apoptosis. In our study, pre-B ALL cases with increased HDM4 or HDM2 expression lacked p53 gene mutation, supporting a role in the negative regulation of p53 function.
Patients with Ph+ pre-B ALL had a worse prognosis than those with Ph− ALL, in agreement with the findings of other studies. The BCR/ABL oncoprotein, the gene product of the t(9;22)(q34;q11) cytogenetically recognized as the Ph, is responsible for transformation of hematopoietic stem cells through deregulated tyrosine kinase activity. Such activity is essential for recruitment and activation of multiple pathways that transduce signals, leading to enhanced proliferative potential and survival. Recently, Trotta et al30 reported that the BCR/ABL oncoprotein can activate HDM2 mRNA translation via the human La antigen and enhance HDM2-mediated inactivation/degradation of p53. In our study, HDM2 and HDM4 showed no correlation with Ph status.
Also, there is evidence that alterations of the HDM4 locus, located at chromosome 1q32, may contribute to the development of some human cancers, including breast carcinoma and glioma. Mitelman et al31 reported that 42 of 647 (6.5%) patients with ALL and chromosome 1 abnormalities involved the 1q32 locus, including 25 (3.9%) with duplications of 1q32. These data suggest that this locus may play a role in the pathogenesis of ALL. Interestingly, none of the 55 pre-B ALL cases in the present study showed a chromosomal abnormality involving chromosome 1q32 as assessed by conventional cytogenetics.
The recent discovery that the HDM4 locus is a regulatory region of both the HDM2 and p53 pathways suggests that tumors with inactivation of the HDM4 locus harbor fewer p53 gene mutations than tumors with an intact HDM2 gene. As p53 is directly targeted in approximately 50% of all human malignancies, it has been postulated that other mechanisms are required to shut down the p53 pathway in tumors with wild-type p53.28, 29
In conclusion, expression of HDM4 and HDM2, which are critical regulators of cell cycle progression, are frequent and abnormal in pre-B ALL. The overall detection rate of HDM4 expression in pre-B ALL cases in this study, approximately 80%, indicates that HDM4 is a potential therapeutic target.
References
Copelan EA, McGuire EA . The biology and treatment of acute lymphoblastic leukemia in adults. Blood 1995;85:1151–1168.
Laport GF, Larson RA . Treatment of adult acute lymphoblastic leukemia. Semin Oncol 1997;24:70–82.
Kantarjian H, Thomas D, O'Brien S, et al. Long-term follow-up results of hyperfractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone (Hyper-CVAD), a dose-intensive regimen, in adult acute lymphocytic leukemia. Cancer 2004;101:2788–2801.
Iwakuma T, Lozano G . MDM2, an introduction. Mol Cancer Res 2003;1:993–1000.
Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J 1996;15:5349–5357.
Marine JC, Jochemsen AG . Mdmx as an essential regulator of p53 activity. Biochem Biophys Res Commun 2005;331:750–760.
Migliorini D, Lazzerini Denchi E, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 2002;22:5527–5538.
Montes de Oca Luna R, Wagner DS, Lozano G . Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203–206.
Jones SN, Roe AE, Donehower LA, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995;378:206–208.
Sharp DA, Kratowicz SA, Sank MJ, et al. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 1999;274:38189–38196.
Stad R, Little NA, Xirodimas DP, et al. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep 2001;2:1029–1034.
Gu J, Kawai H, Nie L, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 2002;277:19251–19254.
Danovi D, Meulmeester E, Pasini D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 2004;24:5835–5843.
Agirre X, Novo FJ, Calasanz MJ, et al. TP53 is frequently altered by methylation, mutation, and/or deletion in acute lymphoblastic leukaemia. Mol Carcinog 2003;38:201–208.
Garcia-Manero G, Bueso-Ramos C, Daniel J, et al. DNA methylation patterns at relapse in adult acute lymphocytic leukemia. Clin Cancer Res 2002;8:1897–1903.
Imamura J, Miyoshi I, Koeffler HP . p53 in hematologic malignancies. Blood 1994;84:2412–2421.
Kantarjian HM, O'Brien S, Smith TL, et al. Results of treatment with hyper-CVAD, a dose-intensive regimen, in adult acute lymphocytic leukemia. J Clin Oncol 2000;18:547–561.
Bueso-Ramos C, Xu Y, McDonnell TJ, et al. Protein expression of a triad of frequently methylated genes, p73, p57Kip2, and p15, has prognostic value in adult acute lymphocytic leukemia independently of its methylation status. J Clin Oncol 2005;23:3932–3939.
Bueso-Ramos CE, Rocha FC, Shishodia S, et al. Expression of constitutively active nuclear-kappa B RelA transcription factor in blasts of acute myeloid leukemia. Hum Pathol 2004;35:246–253.
de Graaf P, Little NA, Ramos YF, et al. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem 2003;278:38315–38324.
Rallapalli R, Strachan G, Cho B, et al. A novel MDMX transcript expressed in a variety of transformed cell lines encodes a truncated protein with potent p53 repressive activity. J Biol Chem 1999;274:8299–8308.
Giglio S, Mancini F, Gentiletti F, et al. Identification of an aberrantly spliced form of HDMX in human tumors: a new mechanism for HDM2 stabilization. Cancer Res 2005;65:9687–9694.
Bartel F, Schulz J, Bohnke A, et al. Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int J Cancer 2005;117:469–475.
Kojima K, Konopleva M, Samudio IJ, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood 2005;106:3150–3159.
Coll-Mulet L, Iglesias-Serret D, Santidrian AF, et al. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood 2006;107:4109–4114.
Hu B, Gilkes DM, Farooqi B, et al. MDMX overexpression prevents P53 activation by the MDM2 inhibitor nutlin. J Biol Chem 2006;281:33030–33035.
Patton JT, Mayo LD, Singhi AD, et al. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res 2006;66:3169–3176.
Ramos YF, Stad R, Attema J, et al. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res 2001;61:1839–1842.
Riemenschneider MJ, Buschges R, Wolter M, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 1999;59:6091–6096.
Trotta R, Vignudelli T, Candini O, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell 2003;3:145–160.
Mitelman F . Catalog of Chromosome Aberrations in Cancer, 5th edn. Wiley-Liss: New York, 1994.
Acknowledgements
Parts of this work were presented at the US and Canadian Academy of Pathology Annual Meeting, San Antonio, TX, USA, February 26–March 4, 2005. Dr X Han is the recipient of the Research Fellowship awarded by the Division of Pathology and Laboratory Medicine, The University of Texas, MD Anderson Cancer Center. We are grateful to Donald R Norwood, LaKisha Rodgers, and Ana Martinez for help in preparation of the manuscript and to Remigio Lopez, Vilmos Thomazy, and Shawn Brisbay for excellent technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Han, X., Garcia-Manero, G., McDonnell, T. et al. HDM4 (HDMX) is widely expressed in adult pre-B acute lymphoblastic leukemia and is a potential therapeutic target. Mod Pathol 20, 54–62 (2007). https://doi.org/10.1038/modpathol.3800727
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800727
Keywords
This article is cited by
-
Gene and protein analysis reveals that p53 pathway is functionally inactivated in cytogenetically normal Acute Myeloid Leukemia and Acute Promyelocytic Leukemia
BMC Medical Genomics (2017)
-
MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer
Journal of Hematology & Oncology (2017)
-
MDMX is a prognostic factor for non-small cell lung cancer and regulates its sensitivity to cisplatin
Cellular Oncology (2017)
-
In vitro and in silico studies of MDM2/MDMX isoforms predict Nutlin-3A sensitivity in well/de-differentiated liposarcomas
Laboratory Investigation (2013)
-
Chromosome instability and deregulated proliferation: an unavoidable duo
Cellular and Molecular Life Sciences (2012)