Abstract
Hepatocellular carcinoma is a highly lethal cancer that typically has poor prognosis. Prognostic markers can help in its clinical management and in understanding the biology of poor prognosis. Through an earlier gene expression study, we identified N-Myc downregulated gene 1 (NDRG1) to be significantly highly expressed in hepatocellular carcinoma compared to nontumor liver. As NDRG1 is a differentiation-related gene with putative metastasis suppressor activity, we investigated the clinical significance of its overexpression. Quantitative real-time polymerase chain reaction using an independent set of patient samples confirmed the significant overexpression of NDRG1 in hepatocellular carcinoma compared to nontumor liver samples (P<0.001). Additionally, high levels of NDRG1 transcript correlated with shorter overall survival (P<0.001), late tumor stage (P=0.001), vascular invasion (P=0.003), large tumor size (P=0.011), and high Edmondson-Steiner histological grade (P=0.005). Using immunohistochemistry, NDRG1 protein was found to be significantly overexpressed in hepatocellular carcinoma samples compared to nontumor liver or cirrhotic and benign liver lesions (P<0.001). Among the hepatocellular carcinoma samples, those which are moderately and poorly differentiated express higher levels of NDRG1 protein than those which are well-differentiated (P<0.005). Additionally, hepatocellular carcinomas with vascular invasion also express elevated levels of NDRG1 protein compared to those without vascular invasion (significant at P<0.005). Our results suggest NDRG1 to be a likely tumor marker for hepatocellular carcinoma, the overexpression of which is correlated with tumor differentiation, vascular invasion, and overall survival. Its significantly elevated expression in hepatocellular carcinoma could be a useful indicator of tumor aggressiveness and therefore patient prognosis.
Similar content being viewed by others
Main
N-Myc downstream regulated gene 1 (NDRG1; also known as Drg-1, Cap43, RTP) is a ubiquitously expressed member of the NDRG family. Regulation of its expression is extremely complex since diverse physiological and pathological conditions (such as hypoxia, cellular differentiation, heavy metal, N-myc, neoplasia) modulate not only NDRG1 transcription, but also its mRNA stability and translation.1 Its diverse regulatory mechanisms and multiple cellular localizations suggest functional involvement in multiple cellular processes. NDRG1 has been proposed to be a signaling protein shuttling between the cytoplasm and nucleus to govern important functions such as growth arrest and cellular differentiation.2 Most notably, NDRG1 is strongly upregulated by hypoxia,3, 4, 5 a condition prevalent in solid tumors. Indeed, the overexpression of NDRG1 has been observed in various types of solid tumors, such as those of the lung, brain, skin, kidney, liver, and breast,4, 6 and could possibly offer an adaptive and protective mechanism for tumor cells to continue growing under anoxic environments. The hypoxic regulation of NDRG1 can occur via hypoxia-inducible factor 1 (HIF-1) dependent as well as HIF-1 independent pathways; its expression is therefore not limited to tissues expressing HIF-1. Additionally, since NDRG1 protein is more stable than HIF-1 protein, NDRG1 has been touted as a better tumor marker than HIF-1.4
Overexpression of NDRG1 in hepatocellular carcinoma has been consistently observed by our group7 and others,8, 9 and appears to be related to hepatocarcinogenesis regardless of the underlying cause (whether or not it is related to chronic hepatitis B or C, the major risk factors of hepatocellular carcinoma). Indeed, the nonstructural protein NS5A of the hepatitis C virus was identified to interact with NDRG1,10 implying a possible role of NDRG1 in the signaling pathways leading to hepatocellular carcinoma. Notably, NDRG1 expression appears to be associated with hepatocellular carcinoma tumor differentiation grade.8 These observations prompted us to further examine the clinical utility of NDRG1 as a tumor marker, and its possible association with other indicators of patient prognosis.
Materials and methods
Gene Expression Data Analysis
Gene expression data based on our earlier study7 were retrieved from the Stanford Microarray Database according to the following selection criteria: all nonflagged spots with >1.5-fold intensities over local background in either channel, 75% good data, and genes whose log (base 2) of red/green normalized ratio (mean) was greater than 3-fold for at least four arrays. The resulting data for 4841 cDNA clones in 75 liver tumor and 72 nontumor liver tissues were uploaded onto the web-based microarray data analysis program GABRIEL (Genetic Analysis By Rules Incorporating Expert Logic; http://gabriel.stanford.edu) for further analysis.
GABRIEL is a rule-based computer program designed to apply domain-specific and procedural knowledge systematically for the analysis and interpretation of data from DNA microarrays.11 In the Chen et al study,7 both the tumor samples (T) and nontumor samples (N) were compared to a pooled universal cell line reference (U) such that expression ratios were represented as log2(T/U) and log2 (N/U), respectively. In order to derive more biologically meaningful data, we rescaled the data set such that the relative T vs N expression ratios (log2(T/N)) is calculated by log2(T/U)−log2(N/U). If a hepatocellular carcinoma sample did not have a corresponding nontumor sample, the global mean of the nontumor gene expression ratios were used. We used the t-score pattern based rule to identify genes which are significantly differentially expressed between tumor and nontumor liver tissues; the t-score (average/standard deviation) allows variability among samples to be taken into consideration during the calculation.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed as described.12, 13 Briefly, quantification was performed using the ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA, USA). Human 18s rRNA primers and probe reagents (Pre-Developed TaqMan Assay Reagents, Applied Biosystems, Foster City, CA, USA) were used as the normalization control for subsequent multiplexed reactions. Primers and probe for NDRG1 were NDRG1_F, 5′-GCC GCC TCC AAG ATC TCA-3′; NDRG1_R, 5′-ACG TTA CTC TGC ATT TCT TCC TTC-3′; and NDRG1_P, 5′-CCC AAG CTC TGC CGG-3′. Transcript quantification was performed in at least duplicates for every sample. The relative amount of NDRG1, which had been normalized with control 18s for RNA amount variation and relative to calibrator, was presented as the relative fold change in log base 2). The calibrator control was the sample with baseline level of the transcript within the normal liver sample series. The NDRG1 level was expressed as ΔΔCt, where  test sample.
test sample.
Statistical Methods
The gene expression data of NDRG1 obtained by quantitative real-time PCR data were considered continuous variables. The data were modeled as categorical variable in the Kaplan–Meier and Cox regression analyses. The Youden index (sensitivity+specificity−1)14 was used to determine the optimal cutoff point of NDRG1 expression for the prediction of overall survival. The Youden index was used to maximize the sensitivity (true positive fraction) and specificity (1−false positive fraction) of the prediction simultaneously. The association of NDRG1 transcript level and clinicopathological features with overall and disease-free survival was examined by multivariate Cox proportional hazards regression using the forward stepwise selection procedure. The NDRG1 transcript levels and their associations with clinicopathological variables were assessed by Fisher's exact test. The statistical analyses were aided by the SPSS version 12.0 software package (SPSS Inc., Chicago, IL, USA). Differences were considered significant when P-value is less than 0.05.
Immunohistochemistry
Immunohistochemistry was performed using microwave heat induced epitope retrieval in citrate buffer as previously described.15 In order to avoid interference from endogenous biotin, a biotin-free method, EnVision™ was used for amplification of the signal (Dako, Carpinteria, CA, USA). Rabbit polyclonal affinity-purified antipeptide antisera against NDRG1 were generated by Applied Genomics Inc. (Sunnyvale, CA, USA).
We used four tissue microarrays for this study, which consisted of archived tissues retrieved from surgical pathology files. In total, there were 23 normal livers and 120 malignant hepatocellular carcinomas, among other control tissues. Tissue arrays were constructed as previously described,16, 17 with core sizes ranging from 0.6 to 2 mm on different arrays. Arrays were scored using a four-tier scale: 0—negative (no staining), 1—equivocal, 2—weak positive (weak to moderate intensity in any percentage of tumor cells), and 3—strong positive (strong intensity in at least 50% of tumor cells).
Results
NDRG1 Transcript is Significantly Overexpressed in Hepatocellular Carcinoma Compared to Nontumor Liver
In an earlier gene expression study of hepatocellular carcinoma, NDRG1 was among the ∼1600 genes observed to be differentially expressed in hepatocellular carcinoma compared to nontumor liver tissue.7 We used a more stringent and biologically relevant analytical approach to verify this observation: first, we rescaled the data set such that the relative tumor (T) vs nontumor (N) expression ratios (log2(T/N)) is calculated by log2(T/U)−log2(N/U), where U is the pooled cell line reference used in the microarray study. If a hepatocellular carcinoma sample did not have a corresponding nontumor sample, the global mean of the nontumor gene expression ratios were used. We then used a t-score pattern based algorithm to find genes satisfying the pattern of being upregulated in tumor compared to nontumor, using a t-score greater than 2. The t-score is a measure of variability (average/standard deviation), and thus provides an indication of the consistency of the expression values across all samples. NDRG1 (t-score 4.697) is among the 1616 genes satisfying this condition; the high t-score implies consistent upregulation of the gene across tumor samples compared to nontumors (Figure 1a).
(a) Relative expression of NDRG1 in hepatocellular carcinoma tumor compared to nontumor liver. Of the total of 75 patients, 57 are matched hepatocellular carcinoma and nontumor liver samples. In the study by Chen et al,7 both the tumor samples (T) and normal tissue samples (N) were compared to a pooled universal cell-line reference (U) such that expression ratios were represented as log2 (T/U) and log2(N/U), respectively. The relative T vs N expression ratios (log2(T/N)) depicted here is calculated by log2(T/U)−log2(N/U). If a hepatocellular carcinoma sample did not have a corresponding nontumor sample, the global mean of the nontumor gene expression ratios were used. The relative abundance of transcripts of each gene is represented by the color scale at the bottom right corner. (b) Quantitative real-time PCR measurement of NDRG1 transcript confirmed its significant overexpression in tumor compared to adjacent nontumor liver and normal liver.
We used quantitative real-time PCR assays to validate the observed upregulation of NDRG1 in hepatocellular carcinoma using an independent set of patient samples consisting of 10 normal liver tissues, 59 hepatocellular carcinoma tissues, and 59 matched adjacent nontumor liver tissues. NDRG1 was found to be expressed at significantly higher levels in hepatocellular carcinomas compared to adjacent nontumors (P<0.001) and normal livers (P<0.001). There was no significant difference between nontumors and normal livers (P=0.880) (Figure 1b).
Overexpression of NDRG1 Transcript in Hepatocellular Carcinoma Correlates with Poor Patient Survival
We investigated the association between NDRG1 expression and patient survival using the Kaplan–Meier analysis. Patients were segregated into low or high NDRG1 level groups, using the Youden index to determine the optimal cutoff value, which was 6.52 for risk segregation. There were 18 deaths (37.5%) within the group of 48 patients with low NDRG1 levels (range: 2.13–6.50), and 9 deaths (81.8%) within the group of 11 patients with high NDRG1 levels (range: 6.54–8.54). Patients with high NDRG1 levels had a significantly shorter overall survival than those with low levels (log-rank test, P<0.001) (Figure 2).
Cox regression analysis was used to compare the NDRG1 levels with clinicopathological features of survival prediction. By univariate Cox regression analysis, high NDRG1 level, vascular invasion, and late tumor stage were significantly associated with poor overall survival (Table 1). By multivariate Cox regression analysis, only high NDRG1 level and vascular invasion were independent prognostic factors for overall survival (Table 1).
Overexpression of NDRG1 Transcript is Associated with Indicators of Poor Prognosis in Hepatocellular Carcinoma
As NDRG1 levels were observed to correlate with overall patient survival, we further examined the associations of NDRG1 transcript levels in tumor tissues with clinicopathological features indicative of poor prognosis in hepatocellular carcinoma. We found that upregulation of NDRG1 transcript was significantly associated with late tumor stage (P=0.001), large tumor size (P=0.018), vascular invasion (P=0.004), and high Edmondson-Steiner histological grade (P=0.012) (Table 2).
There was no association between NDRG1 transcript level and gender, age, HBsAg positivity, liver cirrhosis, or tumor encapsulation.
NDRG1 Protein Overexpression in Hepatocellular Carcinoma Correlates with Tumor Differentiation Grade and Vascular Invasion
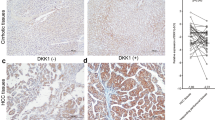
Immunohistochemistry of liver tissue arrays further confirmed the overexpression of NDRG1 protein in liver tumor tissues compared to nontumor liver (P<0.001), and to cirrhotic liver and other benign liver tumors (such as focal nodular hyperplasia, hemangioma, and adenoma) (P<0.001) (Table 3A; Figure 3). Expression of NDRG1 was observed in both the membrane and cytoplasm of malignant hepatocytes, and varied in intensity and extent of staining in different tumors.
Immunohistochemistry of NDRG1 in (a) nontumor liver; (b) cirrhotic liver; (c) well-differentiated hepatocellular carcinoma; (d) moderately differentiated hepatocellular carcinoma; (e) poorly differentiated hepatocellular carcinoma; and (f) hepatocellular carcinoma with vascular invasion. Photos were taken at × 20 magnification.
NDRG1 was initially discovered as a differentiation-related gene, and has been reported to be associated with tumor differentiation in hepatocellular carcinoma.8 We further investigated this association in our patient cohort, and observed that NDRG1 expression was significantly higher in poorly differentiated hepatocellular carcinoma than in well- and moderately differentiated hepatocellular carcinomas combined (P<0.005) (Table 3B; Figure 3). Moreover, well- and moderately differentiated hepatocellular carcinomas tend to show membrane staining, whereas poorly differentiated hepatocellular carcinomas tend to show both membrane and cytoplasmic staining. There was no statistical significance between cellular localization of NDRG1 and tumor differentiation grade.
Of particular clinical importance is the observation that NDRG1 expression correlates with the presence of vascular invasion at both the transcript (Table 2) and protein levels (P<0.005) (Table 3C; Figure 3). As vascular invasion is a strong prognostic indicator of poor patient survival, the overexpression of NDRG1 may contribute towards the aggressiveness of this group of tumors.
No significant association was observed between NDRG1 expression/localization and other clinicopathologic parameters, such as α-fetoprotein level, tumor size, or tumor recurrence.
Discussion
Hepatocellular carcinoma, the primary adult liver malignancy, is the third leading cause of cancer deaths worldwide.18, 19 The majority (70–85%) of hepatocellular carcinoma cases arise from chronic infections with hepatitis B virus or hepatitis C virus, with other nonviral causes playing much lesser roles.20, 21 Hepatocellular carcinoma is particularly challenging to treat because patients do not have overt symptoms until the disease is in its advanced stages, making treatment difficult and ineffective. In most cases, curative therapy can be achieved only by surgical hepatic resection; even with surgical resection, the 5-year survival rate is poor (about 35%). Clinically, high tumor differentiation grade and the presence of vascular invasion are significant independent predictors of patient survival.22, 23
An earlier study by our group generated gene expression profiles of hepatocellular carcinoma in order to increase our understanding of the molecular pathophysiology of hepatocellular carcinoma, as well as to identify genetic fingerprints with diagnostic or prognostic significance.7 We reanalyzed these gene expression profiles using GABRIEL in order to extract biologically and clinically meaningful results, and identified NDRG1 as one of the most significantly upregulated genes in hepatocellular carcinoma when compared to nontumor liver. The significantly elevated expression of NDRG1 in hepatocellular carcinoma was further validated by quantitative real-time PCR in an independent set of patients. Additionally, high levels of NDRG1 transcript was found to be significantly correlated with short overall survival, late tumor stage, large tumor size, the presence of vascular invasion, and high histological grade.
Most importantly, the overexpression of NDRG1 transcript in hepatocellular carcinoma was reflected at the protein level (as detected by immunohistochemistry in our tissue arrays), suggesting utility as a histologic tumor marker of hepatocellular carcinoma. Its ability to differentiate between hepatocellular carcinoma tissues and cirrhotic liver further adds to its clinical value as a diagnostic marker. Additionally, the expression of NDRG1 protein was strongly correlated with the tumor differentiation grade, and implies some functional significance of NDRG1 in tumor progression. In agreement with our observation, Wamunyokoli et al24 reported that the expression of NDRG1 is dependent of tumor grade in esophageal cancer.
Hepatocellular carcinoma is a highly vascular and proliferative tumor. As vascular invasion is an important step towards tumor spread and metastasis, it is an independent prognostic indicator of recurrence and long-term survival of hepatocellular carcinoma patients.25, 26 Many factors modulate the propensity of malignant tumors for vascular invasion, of which altered cell–cell and cell–matrix interactions play a critical role.27, 28, 29 Additionally, factors involved in angiogenesis (eg vascular endothelial growth factor receptor),25 cell differentiation and growth arrest (eg annexin A10),30 tumor suppression (eg p53),31, 32 and modifications of the extracellular matrix (eg matrix metalloproteinases)33, 34, 35 play equally important roles in vascular invasion and tumor spread. Our observation that NDRG1 overexpression in hepatocellular carcinoma correlates significantly with vascular invasion suggests a role of NDRG1 in the spread of this tumor, and its role as a prognostic indicator of hepatocellular carcinoma patient survival. However, a recent study reported that NDRG1 suppresses in vitro invasion of hepatoma cells,36 which contradicts our clinical observations.
The overexpression of NDRG1 in various types of cancers has been reported.3, 4 Functionally, NDRG1 has been implicated as a metastasis suppressor. In prostate cancer, downregulation of NDRG1 was reported to correlate with advancement and metastatic progression of the disease.37 A similar putative metastatic suppressor gene function for NDRG1 has also been suggested in human colon cancer.38, 39, 40 However, another study by Wang et al41 contradicts these earlier studies by showing that the expression of NDRG1 gradually increases during colorectal carcinogenesis and that NDRG1 might be a putative tumor metastasis promoter gene. Possible mutations at both the transcriptional and translational levels may account for these discrepancies in its roles in cancer. Specifically, NDRG1 is reported to be a multiphosphorylated protein;42, 43 the role of phosphorylation is unknown but speculated to be related to the multitude of physiological functions of NDRG1.
In conclusion, NDRG1 is a potential tumor marker of hepatocellular carcinoma, the overexpression of which is also correlated with overall survival, late tumor stage, large tumor size, tumor differentiation grade, and vascular invasion. As patients with high-grade tumor and vascular invasion tend to have poorer prognosis, it is logical that NDRG1 may serve as a strong prognostic indicator of hepatocellular carcinoma. Additionally, the possibility of regulating NDRG1 expression levels via multiple pathways makes it a potential therapeutic target in hepatocellular carcinoma. Full characterization of the biological functions of NDRG1 in human hepatoma cell lines is underway to investigate the complex roles it may play in the process of hepatocarcinogenesis, and how it may be exploited for the clinical management of hepatocellular carcinoma.
References
Lachat P, Shaw P, Gebhard S, et al. Expression of NDRG1, a differentiation-related gene, in human tissues. Histochem Cell Biol 2002;118:399–408.
Kalaydjieva L, Gresham D, Gooding R, et al. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet 2000;67:47–58.
Cangul H, Salnikow K, Yee H, et al. Enhanced overexpression of an HIF-1/hypoxia-related protein in cancer cells. Environ Health Perspect 2002;110:783–788.
Cangul H . Hypoxia upregulates the expression of the NDRG1 gene leading to its overexpression in various human cancers. BMC Genet 2004;5:27, doi:10.1186/1471-2156/5/27.
Masuda K, Ono M, Okamoto M, et al. Downregulation of Cap43 gene by von Hippel-Lindau tumor suppressor protein in human renal cancer cells. Int J Cancer 2003;105:803–810.
Cangul H, Salnikow K, Yee H, et al. Enhanced expression of a novel protein in human cancer cells: a potential aid to cancer diagnosis. Cell Biol Toxicol 2002;18:87–96.
Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell 2002;13:1929–1939.
He J, Zhou R . Correlation of NDRG1 gene with liver tissue differentiation and hepatocarcinogenesis. Beijing Da Xue Xue Bao 2003;35:471–475.
Kurokawa Y, Matoba R, Takemasa I, et al. Molecular features of non-B, non-C hepatocellular carcinoma: a PCR-array gene expression profiling study. J Hepatol 2003;39:1004–1012.
Ahn J, Chung K-S, Kim D-U, et al. Systematic identification of hepatocellular proteins interacting with NS5A of the hepatitis C virus. J Biochem Mol Biol 2004;37:741–748.
Pan K-H, Lih C-J, Cohen SN . Analysis of DNA microarrays using algorithms that employ rule-based expert knowledge. Proc Natl Acad Sci 2002;99:2118–2123.
Cheung ST, Ho JC, Leung KL, et al. Transcript AA454543 is a novel prognostic marker for hepato-cellular carcinoma after curative partial hepatectomy. Neoplasia 2005;7:91–98.
Cheung ST, Leung KL, Ip YC, et al. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res 2005;11 (2 Part 1):551–556.
Youden WJ . Index for rating diagnostic tests. Cancer 1950;3:32–35.
Higgins JP, Warnke RA . CD30 expression is common in mediastinal large B-cell lymphoma. Am J Clin Pathol 1999;112:241–247.
Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–847.
Van De Rijn M, Perou CM, Tibshirani R, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol 2002;161:1991–1996.
Block TM, Mehta AS, Fimmel CJ, et al. Molecular viral oncology of hepatocellular carcinoma. Oncogene 2003;22:5093–5107.
El-Serag HB . Hepatocellular carcinoma. J Clin Gastroenterol 2002;35 (Suppl 2):S72–S78.
Suriawinata A, Xu R . An update on the molecular genetics of hepatocellular carcinoma. Sem Liver Dis 2004;24:77–88.
Szabo E, Paska C, Novak PK, et al. Similarities and differences in hepatitis B and C virus induced hepatocarcinogenesis. Pathol Oncol Res 2004;10:5–11.
Kondo K, Chijiiwa K, Makino I, et al. Risk factors for early death after liver resection in patients with solitary hepatocellular carcinoma. J Hepatob Pancreat Surg 2005;12:399–404.
Qin LX, Tang ZY . The prognostic significance of clinical and pathological features in hepatocellular carcinoma. World J Gastroenterol 2002;8:193–199.
Wamunyokoli F, Hendricks D, Parker I . N-Myc downstream regulated gene 1 (NDRG1) expression and localization is tumor grade dependent in oesophageal cancer. Proceedings of the 9th Biochemical Society of Kenya Annual Symposium. African J Biotechnol 2004;3 (Abstract).
Dhar DK, Naora H, Yamanoi A, et al. Requisite role of VEGF receptors in angiogenesis of hepatocellular carcinoma: a comparison with angiopoietin/Tie pathway. Anticancer Res 2002;22:379–386.
Yeh C-N, Chen M-F, Lee W-CH, et al. Prognostic factors of hepatic resection for hepatocellular carcinomas with cirrhosis: univariate and multivariate analysis. J Surg Oncol 2002;81:195–202.
Albelda SM, Buck CA . Integrins and other cell adhesion molecules. FASEB J 1990;4:2868–2880.
Albelda SM . Biology of disease. Role of integrins and other cell adhesion molecules in tumor progression and metastasis. Lab Invest 1993;68:4–17.
Mathew J, Hines JE, Obafunwa JO, et al. CD44 is expressed in hepatocellular carcinomas showing vascular invasion. J Pathol 1996;179:74–79.
Liu SH, Lin CY, Peng SY, et al. Down-regulation of annexin A10 in hepatocellular carcinoma is associated with vascular invasion, early recurrence, and poor prognosis in synergy with p53 mutation. Am J Pathol 2002;160:1831–1837.
Hsu HC, Tseng HJ, Lai OL, et al. Expression of p53 gene in 184 unifocal hepatocellular carcinomas (HCC): association with tumor growth and invasiveness. Cancer Res 1993;53:4691–4694.
Hsu HC, Peng SY, Lai PL, et al. Mutations of p53 gene in hepatocellular carcinoma (HCC) correlate with tumor progression and patient prognosis: a study of 138 patients with unifocal HCC. Int J Oncol 1994;4:1341–1347.
Liotta LA, Tryggvason K, Garbisa S, et al. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature 1980;6:67–68.
Arri S, Mise M, Harada T, et al. Overexpression of matrix metalloproteinase 9 gene in hepatocellular carcinoma with invasive potential. Hepatology 1996;24:316–322.
Murakami K, Sakukawa R, Ikeda T, et al. Invasiveness of hepatocellular carcinoma cell lines: contribution of membrane-type 1 matrix metalloproteinase. Neoplasia 1999;1:424–430.
Hosoi F, Oie S, Maruyama Y, et al. Expression of a metastasis suppressor gene, Cap43/NDRG1/Drg-1 through hepatocyte nuclear factors in human hepatocellular carcinoma cells. Proceedings of the 96th Annual Meeting of the American Association for Cancer Research; 16–20 April 2005; Anaheim/Orange County, USA. Abstract No. 328.
Bandyopadhyay S, Pai SK, Gross SC, et al. The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Res 2003;63:1731–1736.
van Belzen N, Dinjens WN, Diesveld MP, et al. A novel gene which is up-regulated during colon epithelial cell differentiation and down-regulated in colorectal neoplasms. Lab Invest 1997;77:85–92.
van Belzen N, Dinjens WN, Eussen BH, et al. Expression of differentiation-related genes in colorectal cancer: possible implications for prognosis. Histol Histopathol 1998;13:1233–1242.
Guan RJ, Ford HL, Fu Y, et al. Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Res 2000;60:749–755.
Wang Z, Wang F, Wang W-Q, et al. Correlation of N-myc downstream-regulated gene 1 overexpression with progressive growth of colorectal neoplasm. World J Gastroenterol 2004;10:550–554.
Murray JT, Campbell DG, Morrice N, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J 2004;384:477–488.
Agarwala KL, Kokame K, Kato H, et al. Phosphorylation of RTP, an ER stress-responsive cytoplasmic protein. Biochem Biophys Res Comm 2000;272:641–647.
Acknowledgements
We are grateful to Dr Stanley Cohen and Kevin Pan from the Department of Genetics at Stanford University for help with GABRIEL analysis. This work was supported by grants to the Asian Liver Center at Stanford University from the HM Lui and CJ Huang Foundations (to M-S Chua, V Mason and S So); and by the Seed Funding Program of the University of Hong Kong (to ST Cheung and ST Fan).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chua, MS., Sun, H., Cheung, S. et al. Overexpression of NDRG1 is an indicator of poor prognosis in hepatocellular carcinoma. Mod Pathol 20, 76–83 (2007). https://doi.org/10.1038/modpathol.3800711
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800711
Keywords
This article is cited by
-
JUN-induced super-enhancer RNA forms R-loop to promote nasopharyngeal carcinoma metastasis
Cell Death & Disease (2023)
-
NDRG1 is induced by antigen-receptor signaling but dispensable for B and T cell self-tolerance
Communications Biology (2022)
-
Biological aspects in controlling angiogenesis: current progress
Cellular and Molecular Life Sciences (2022)
-
The androgen receptor expression and its activity have different relationships with prognosis in hepatocellular carcinoma
Scientific Reports (2020)
-
Upregulation of NDRG1 predicts poor outcome and facilitates disease progression by influencing the EMT process in bladder cancer
Scientific Reports (2019)