Abstract
RHAMM, a member of the microtubule-associated protein family that interacts with the mitogen-activated protein kinase pathway, is associated with tumor progression, aggressive disease and shortened survival in several tumor types. This study aimed to determine the prognostic value of RHAMM in colorectal cancer (CRC). A series of 1420 unselected, nonconsecutive CRC resections were subdivided into three groups: (1) DNA mismatch repair (MMR)-proficient, (2) MLH1 negative and (3) presumed Lynch syndrome. Immunohistochemical analysis of RHAMM expression (0 vs >0%), increasing expression (increasing percentage positivity) and complete expression (100 vs <100%) was performed using tissue microarray technique and the results were correlated with clinicopathological parameters. Fifty-seven tissue samples of normal colonic mucosa were included as a control group. In a univariate analysis increasing and complete expression of RHAMM were associated with higher N stage (P=0.023 and 0.021) and worse survival (P<0.0001) in MMR-proficient CRC. Complete expression of RHAMM was associated with worse survival in presumed Lynch syndrome (P=0.016). In MLH1-negative CRC there was no association between RHAMM expression and the clinicopathological features. In a multivariate analysis, increasing RHAMM expression was an independent adverse prognostic factor in MMR-proficient CRC (P<0.0001) and complete expression in MMR-proficient CRC and presumed Lynch syndrome (P<0.0001 and P=0.031, respectively). Nuclear pERK expression was associated with increasing RHAMM expression in MMR-proficient CRC (P=0.012) and with complete RHAMM expression in presumed HNPCC (P=0.03). Increasing and complete RHAMM expressions are independent adverse prognostic factors in MMR-proficient CRC and presumed Lynch syndrome.
Similar content being viewed by others
Main
The receptor for hyaluronic acid mediated motility (RHAMM; CD168, intracellular hyaluronic acid binding protein) has a cell surface and intracellular distribution.1 RHAMM binds hyaluronan,2 interacts with both mictotubules and microfilaments,3, 4 localizes to the centrosome maintaining the spindle integrity5 and is suggested to represent a member of the MAP family.3 RHAMM is involved in cell motility and signaling6 as well as oncogenic events.7
The mitogen-activated protein kinase (MAPK) pathway includes several families of signal transduction cascades which mediate information provided by extracellular stimuli.8 The MAPK pathway is considered to be important for cellular growth, development and differentiation and regulates cell proliferation, apoptosis, cell differentiation and tissue development.9 The Raf-MEK-ERK pathway belongs to the MAPK pathways and represents one of the best characterized Ras signaling pathways.10 The molecule ERK is activated by a cascade of phosphorylation events downstream from the ras proto-oncogene8 and plays a role in differentiation, secretion, proliferation and hypertrophy.11 RHAMM binds ERK kinase12 and controls expression levels of ERK.13
There is evidence that RHAMM influences tumor progression and metastasis in different tumor types including pancreatic cancer,14 stomach cancer,15 endometrial carcinomas,16 breast cancer,4, 13, 17 transitional cell carcinomas of urinary bladder,18 aggressive fibromatosis (desmoid tumor),19 lung cancer,20 B-cell malignancies1, 21, 22, 23 and melanoms.24 However, the prognostic significance of RHAMM in colorectal cancer (CRC) is poorly understood. The aim of this study was therefore to determine the prognostic significance of expression, increasing expression and complete expression of RHAMM assessed by means of immunohistochemistry (IHC) in 1420 tissue microarray (TMA) specimens stratified into mismatch repair (MMR) proficient, MLH1 negative and presumed Lynch syndrome/hereditary nonpolyposis colorectal cancer (HNPCC) and to investigate the interaction between RHAMM and pERK.
Materials and methods
Tissue Microarray Construction
A TMA of 1420 unselected, nonconsecutive CRCs was constructed as described previously.25 Formalin-fixed, paraffin-embedded tissue blocks of CRC resections were retrieved from the archives of the Institute of Pathology, University Hospital of Basel, Switzerland, the Institute of Clinical Pathology, Basel, Switzerland and the Institute of Pathology, Stadtspital Triemli, Zürich, Switzerland. One tissue cylinder with a diameter of 0.6 mm was punched from morphologically representative tissue areas of each ‘donor’ tissue block and brought into one recipient paraffin block (3 × 2.5 cm) using a homemade semiautomated tissue arrayer. Failure of analysis including missing samples or fractions containing only a few tumor cells was related to tissue microarray technology.
Clinicopathological Data and Tumors
CRC resections were subdivided into three CRC subsets: (1) DNA MMR-proficient (expressing MLH1, MSH2 and MSH6), (2) MLH1 negative and (3) presumed Lynch syndrome/HNPCC (MSH2 and/or MSH6 negative at any age or MLH1 negative and <55 years of age). These immunohistochemical groupings presented a good fit to the known clinicopathological features associated with these groups of CRC. While a small proportion of presumed sporadic MSI-H and HNPCC cases may have been incorrectly assigned, the overall findings are likely to be valid in view of the large numbers of samples and the good fit with clinicopathological features.
One pathologist (L.Te.) systematically re-evaluated the clinicopathological data with respect to CRCs. The clinicopathological data of the different CRC subsets are summarized in Table 1. Any disagreement between the clinicopathological features and numbers of available tissue punches shown in Table 1 is due to missing clinicopathological data.
Immunohistochemistry of TMA
Sections (4 μm) of TMA blocks were transferred to an adhesive-coated slide system (Instrumedics Inc., Hackensack, NJ, USA) to facilitate the transfer of tissue microarray sections to slides and to minimize tissue loss. Standard indirect immunoperoxidase procedures were used for immunohistochemistry (ABC-Elite, Vector Laboratories, Burlingame, CA, USA). 1420 CRCs and 57 normal colonic mucosa samples were immunostained for RHAMM (clone 2D6; dilution 1:25, Novocastra, UK), MLH1 (clone MLH-1; dilution 1:100; BD Biosciences Pharmingen, San Jose, CA, USA), MSH2 (clone MSH-2; dilution 1:200; BD Biosciences Pharmingen, San Jose, CA), MSH6 (clone 44; dilution 1:400; BD Biosciences Pharmingen, San Jose, CA). After dewaxing and rehydration in dH2O, sections for immunostaining were subjected to heat antigen retrieval in a microwave oven (1200 W, 15 min) in 1 mM EDTA buffer pH 9.0 for RHAMM, 0.001 mol/l ethylenediaminetetraacetic acid, pH 8.0 for MLH1 and MSH2 and 0.01 mol/l citrate buffer pH 6.0 for MSH6. Endogenous peroxidase activity was blocked using 0.5% H2O2. After transfer to a humidified chamber, the sections were incubated with 10% normal goat serum (Dako Cytomation) for 20 min and incubated with primary antibody at 4°C overnight for hMLH1, hMSH2 and hMSH6 and at room temperature for RHAMM (1 h). Subsequently, the sections were incubated with peroxidase-labeled polymer (K4005, EnVision+ System-HRP(AEC); DakoCytomation) for 30 min at room temperature. For visualization of the antigen, the sections were immersed in 3-amino-9-ethylcarbazole+substrate-chromogen (K4005, EnVision+ System-HRP (AEC); DakoCytomation) for 30 min, and counterstained lightly with Gill's haematoxylin.
RHAMM immunoreactivity was evaluated using the percentage of positive cells ranging from 0 to 100%. RHAMM expression was defined as 0 vs >0%, increasing expression as increasing percentage positivity and complete expression as 100 vs <100%. Normal colonic mucosa was considered as baseline to determine RHAMM expression in CRC and cells were scored as positive when the intensity of baseline expression was clearly exceeded. Immunohistochemistry for MLH1, MSH2 and MSH6 was scored as negative when no staining (0%) was observed and as positive when any immunoreactivity (>0%) was found.
Statistical Analysis
Clinicopathological characteristics across CRC groups were analyzed using the Kruskal–Wallis and χ2 tests. Univariate analysis of 5-year survival rates across CRC groups and according to RHAMM expression and complete expression was carried out using the Kaplan–Meier method and log-rank test. The distribution of RHAMM expression across CRC groups was evaluated using the χ2 test. The association of clinicopathological characteristics and expression, increasing expression and complete expression of RHAMM were performed using univariate regression analysis. To determine whether RHAMM was a prognostic indicator of 5-year survival independent of T stage, N stage, tumor grade and vascular invasion, the Cox-proportional hazard method was used. P-values <0.05 were considered statistically significant. All analyses were carried out using SAS (Version 9.1, The SAS Institute, NC, USA).
Results
Normal Colonic Mucosa
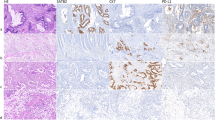
In normal colonic mucosa RHAMM was diffusely but weakly expressed in the cytoplasm of columnar cells of the crypts, but apparently not in the goblet cells (Figure 1a). RHAMM was less strongly expressed as compared to the cancer cell population. A more detailed analysis was limited by the small number of samples and the lack of the surface epithelium in most of the tissue microarray samples.
Distribution of Cytoplasmic RHAMM Expression in the Different CRC Subsets (Table 2)
A different percentage of cytoplasmic RHAMM-positive tumors occurred across MMR proficient (95.7%), MLH1 negative (91.7%) and presumed Lynch syndrome (79.2%) subgroups (P<0.001) (Figure 1b–d).
MMR-Proficient CRC (Table 3)
In a univariate analysis increasing and complete expression of RHAMM was associated with higher N stage (P=0.023 and P=0.021) and worse survival (P<0.0001) in MMR-proficient CRC (Figure 2).
MLH1-Negative CRC (Table 4)
In a univariate analysis there was no association between expression, increasing expression and complete expression of RHAMM and the clinicopathological features including T stage, N stage, tumor grade, vascular invasion and survival.
Presumed Lynch Syndrome (Table 5)
In a univariate analysis RHAMM expression was associated with lower tumor grade (P=0.004) and complete expression with worse survival (P=0.016). Increasing expression of RHAMM was not associated with the clinicopathological parameters.
Multivariate Analysis of Survival
Increasing RHAMM expression was an independent adverse prognostic factor in MMR-proficient CRC (P<0.0001) and complete expression in MMR-proficient CRC and presumed Lynch syndrome (P<0.0001 and P=0.031, respectively).
Association between Cytoplasmic RHAMM and Nuclear pERK (Table 6)
In MMR-proficient CRC increasing RHAMM expression was associated with nuclear pERK expression (P=0.012), whereas complete expression of RHAMM was associated with pERK expression in presumed Lynch syndrome (P=0.03). In MLH1-negative CRC RHAMM was not associated with pERK.
Discussion
In this study, we used TMA technology and IHC on a large number of unselected CRC cases (n=1420), stratifying these according to MMR repair status, and describing cytoplasmic RHAMM-positive tumor cells in every punch sample. This ‘descriptive’ evaluation system has the advantage of defining marker expression (0 vs >0%), increasing expression (percentage of positive tumor cell staining) and complete expression (100 vs <100%) avoiding an ‘interpretative’ and often complex composite scoring system.
In normal colonic mucosa RHAMM was diffusely expressed in the cytoplasm of columar cells in the crypts, but apparently not in the goblet cells. RHAMM expression was weaker and quantitatively less extensive as compared to cancer cells.
Our results point to biological differences between expression (0 vs >0%), increasing expression (increasing percentage positivity) and complete expression (100 vs <100%) of RHAMM in CRC. RHAMM expression was not associated with tumor progression and worse survival in all three CRC subgroups, which can be explained by the fact that RHAMM expression was found in normal colonic mucosa. This suggests that increasing RHAMM expression is needed to induce tumor progression. Indeed, in MMR-proficient CRC increasing and complete expression of RHAMM were correlated with higher N stage and worse survival in a univariate analysis and were independent adverse prognostic factors in a multivariate analysis, whereas in presumed Lynch syndrome complete RHAMM expression was associated with worse survival in univariate and multivariate analysis. These findings fit with the results obtained in a recent study in which using RT-PCR on tissue specimens of patients with CRC RHAMM mRNA levels were higher in tumor tissue when compared to adjacent normal tissue.26
Our findings are also in agreement with previous studies that assessed RHAMM expression and the associations with clinicopathological parameters in different tumor types. In endometrial carcinoma Rein et al16 found a significant correlation between RHAMM expression and lymph node metastasis and Assmann et al4 showed an association between trabecular (trabeculae=single cells in the stroma which appear to be budding off from the main tumor mass) RHAMM expression and worse survival in breast cancer. RHAMM expression was also associated with tumor progression in transitional cell carcinomas of urinary bladder18 and stomach cancer.15
In a recent study, we analyzed the association between expression (0 vs >0%) and increasing expression (increasing percentage of positivity) of nuclear pERK and different clinicopathological parameters including T stage, N stage, tumor grade, tumor budding, vascular invasion and survival in MMR proficient, MLH1 negative and presumed Lynch syndrome.27 Only nuclear pERK expression was associated with tumor budding in the MMR-proficient CRC subgroup, whereas no association was observed between nuclear pERK expression/increasing expression and the clinicopathological features in the MLH1 negative and the presumed Lynch syndrome subgroups. Tumor budding is defined as the presence of isolated single cells or small cell clusters (up to 4) scattered in the stroma at the invasive tumor margin and is established as an adverse prognostic indicator.28, 29, 30, 31, 32, 33 Tumor budding is at least in part driven by the wnt signaling pathway as attested by the fact that nuclear β-catenin accumulates in the nuclei in tumor buds (dedifferentiated cancer cells) at the invasive tumor border.34, 35, 36, 37
In the present study, nuclear pERK expression was correlated with increasing RHAMM expression in MMR proficient (P=0.012) and with complete RHAMM expression in presumed Lynch syndrome (P=0.03), whereas an association was not found in MLH1-negative CRC. This finding leads to the hypothesis that pERK is involved in the mechanism of tumor progression of MMR-proficient CRC and Lynch syndrome by interacting with the wnt signaling pathway and RHAMM: (1) KRAS mutation is found in approximately 35% of unselected CRCs, whereas it is mutated at a particularly low frequency in sporadic MSI-H cancers.38, 39, 40, 41, 42 (2) The molecule ERK, a member of the MAPK pathway, is activated by a cascade of phophorylation events downstream from the ras proto-oncogene.8 (3) Intracellular and cell surface RHAMM isoforms are important for the activation of ERK by PDGF (platelet-derived growth factor) and mutant (activated) RAS, respectively, while intracellular RHAMMv4 overexpression activates ERK.12 (4) ERK interacts with the wnt signaling pathway by inactivating GSK3β which is a part of a complex (with APC and axin) responsible for the degradation of β-catenin.43, 44 (5) Dysregulation of the wnt signaling pathway (activating mutation of β-catenin and inactivating mutation of APC) is more likely to occur in MMR proficient than in MLH1-negative CRC.45, 46 (6) Although oncogenic mutation of β-catenin has been linked with MSI-H CRC, this association is only with Lynch syndrome (and only around 20% of these) and not in MLH1-negative CRC.41, 46
Therefore, pERK may not function in isolation as a prognostic factor in CRC (the same observation was made by by Wang et al13 in breast cancer), but may be implicated in CRC progression through its interactions with the wnt signaling pathway and RHAMM.
In summary, our study has shown that complete expression of RHAMM is an independent adverse prognostic factors in MMR-proficient CRC and presumed Lynch syndrome. Additionally, RHAMM expression is correlated with nuclear pERK in both groups of CRC.
References
Maxwell CA, Keats JJ, Belch AR, et al. Receptor for hyaluronan-mediated motility correlates with centrosome abnormalities in multiple myeloma and maintains mitotic integrity. Cancer Res 2005;65:850–860.
Yang B, Zhang L, Turley EA . Identification of two hyaluronan-binding domains in the hyaluronan receptor RHAMM. J Biol Chem 1993;268:8617–8623.
Assmann V, Jenkinson D, Marshall JF, et al. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J Cell Sci 1999;112 (Part 22):3943–3954.
Assmann V, Gillett CE, Poulsom R, et al. The pattern of expression of the microtubule-binding protein RHAMM/IHABP in mammary carcinoma suggests a role in the invasive behaviour of tumour cells. J Pathol 2001;195:191–196.
Maxwell CA, Keats JJ, Crainie M, et al. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol Biol Cell 2003;14:2262–2276.
Turley EA, Noble PW, Bourguignon LY . Signaling properties of hyaluronan receptors. J Biol Chem 2002;277:4589–4592.
Hall CL, Yang B, Yang X, et al. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell 1995;82:19–26.
Sebolt-Leopold JS, Dudley DT, Herrera R, et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat Med 1999;5:810–816.
Shapiro P . Ras-MAP kinase signaling pathways and control of cell proliferation: relevance to cancer therapy. Crit Rev Clin Lab Sci 2002;39:285–330.
Sebolt-Leopold JS . Development of anticancer drugs targeting the MAP kinase pathway. Oncogene 2000;19:6594–6599.
Marais R, Light Y, Mason C, Paterson H, et al. Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by protein kinase C. Science 1998;280:109–112.
Zhang S, Chang MC, Zylka D, et al. The hyaluronan receptor RHAMM regulates extracellular-regulated kinase. J Biol Chem 1998;273:11342–11348.
Wang C, Thor AD, Moore DH, et al. The overexpression of RHAMM, a hyaluronan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin Cancer Res 1998;4:567–576.
Abetamann V, Kern HF, Elsasser HP . Differential expression of the hyaluronan receptors CD44 and RHAMM in human pancreatic cancer cells. Clin Cancer Res 1996;2:1607–1618.
Li H, Guo L, Li JW, et al. Expression of hyaluronan receptors CD44 and RHAMM in stomach cancers: relevance with tumor progression. Int J Oncol 2000;17:927–932.
Rein DT, Roehrig K, Schondorf T, et al. Expression of the hyaluronan receptor RHAMM in endometrial carcinomas suggests a role in tumour progression and metastasis. J Cancer Res Clin Oncol 2003;129:161–164.
Assmann V, Marshall JF, Fieber C, et al. The human hyaluronan receptor RHAMM is expressed as an intracellular protein in breast cancer cells. J Cell Sci 1998;111 (Part 12):1685–1694.
Kong QY, Liu J, Chen XY, et al. Differential expression patterns of hyaluronan receptors CD44 and RHAMM in transitional cell carcinomas of urinary bladder. Oncol Rep 2003;10:51–55.
Tolg C, Poon R, Fodde R, et al. Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor). Oncogene 2003;22:6873–6882.
Teder P, Bergh J, Heldin P . Functional hyaluronan receptors are expressed on a squamous cell lung carcinoma cell line but not on other lung carcinoma cell lines. Cancer Res 1995;55:3908–3914.
Turley EA, Belch AJ, Poppema S, et al. Expression and function of a receptor for hyaluronan-mediated motility on normal and malignant B lymphocytes. Blood 1993;81:446–453.
Pilarski LM, Masellis-Smith A, Belch AR, et al. RHAMM, a receptor for hyaluronan-mediated motility, on normal human lymphocytes, thymocytes and malignant B cells: a mediator in B cell malignancy? Leuk Lymph 1994;14:363–374.
Masellis-Smith A, Belch AR, Mant MJ, et al. Hyaluronan-dependent motility of B cells and leukemic plasma cells in blood, but not of bone marrow plasma cells, in multiple myeloma: alternate use of receptor for hyaluronan-mediated motility (RHAMM) and CD44. Blood 1996;87:1891–1899.
Ahrens T, Assmann V, Fieber C, et al. CD44 is the principal mediator of hyaluronic-acid-induced melanoma cell proliferation. J Invest Dermatol 2001;116:93–101.
Sauter G, Simon R, Hillan K . Tissue microarrays in drug discovery. Nat Rev Drug Discov 2003;2:962–972.
Line A, Slucka Z, Stengrevics A, et al. Characterisation of tumour-associated antigens in colon cancer. Cancer Immunol Immunother 2002;51:574–582.
Lugli A, Zlobec I, Minoo P, et al. Role of the MAPK and PI3K/AKT pathways downstream molecules pERK and pAKT in colorectal cancer. A tissue-microarray based approach. Hum Pathol 2006, In press.
Hase K, Shatney C, Johnson D, et al. Prognostic value of tumor ‘budding’ in patients with colorectal cancer. Dis Colon Rectum 1993;36:627–635.
Hase K, Shatney CH, Mochizuki H, et al. Long-term results of curative resection of ‘minimally invasive’ colorectal cancer. Dis Colon Rectum 1995;38:19–26.
Shinto E, Mochizuki H, Ueno H, et al. A novel classification of tumour budding in colorectal cancer based on the presence of cytoplasmic pseudo-fragments around budding foci. Histopathology 2005;47:25–31.
Tanaka M, Hashiguchi Y, Ueno H, et al. Tumor budding at the invasive margin can predict patients at high risk of recurrence after curative surgery for stage II, T3 colon cancer. Dis Colon Rectum 2003;46:1054–1059.
Ueno H, Mochizuki H, Hatsuse K, et al. Indicators for treatment strategies of colorectal liver metastases. Ann Surg 2000;231:59–66.
Ueno H, Murphy J, Jass JR, et al. Tumour ‘budding’ as an index to estimate the potential of aggressiveness in rectal cancer. Histopathology 2002;40:127–132.
Adachi Y, Yamamoto H, Itoh F, et al. Clinicopathologic and prognostic significance of matrilysin expression at the invasive front in human colorectal cancers. Int J Cancer 2001;95:290–294.
Pyke C, Salo S, Ralfkiaer E, et al. Laminin-5 is a marker of invading cancer cells in some human carcinomas and is coexpressed with the receptor for urokinase plasminogen activator in budding cancer cells in colon adenocarcinomas. Cancer Res 1995;55:4132–4139.
Brabletz T, Jung A, Hermann K, et al. Nuclear overexpression of the oncoprotein beta-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol Res Pract 1998;194:701–704.
Hlubek F, Jung A, Kotzor N, et al. Expression of the invasion factor laminin gamma2 in colorectal carcinomas is regulated by beta-catenin. Cancer Res 2001;61:8089–8093.
Fujiwara T, Stolker JM, Watanabe T, et al. Accumulated clonal genetic alterations in familial and sporadic colorectal carcinomas with widespread instability in microsatellite sequences. Am J Pathol 1998;153:1063–1078.
Jass JR, Biden KG, Cummings MC, et al. Characterisation of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. J Clin Pathol 1999;52:455–460.
Simms LA, Radford-Smith G, Biden KG, et al. Reciprocal relationship between the tumor suppressors p53 and BAX in primary colorectal cancers. Oncogene 1998;17:2003–2008.
Salahshor S, Kressner U, Pahlman L, et al. Colorectal cancer with and without microsatellite instability involves different genes. Genes Chromosomes Cancer 1999;26:247–252.
Olschwang S, Hamelin R, Laurent-Puig P, et al. Alternative genetic pathways in colorectal carcinogenesis. Proc Natl Acad Sci USA 1997;94:12122–12127.
Ding Q, Xia W, Liu JC, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell 2005;19:159–170.
Yun MS, Kim SE, Jeon SH, et al. Both ERK and Wnt/beta-catenin pathways are involved in Wnt3a-induced proliferation. J Cell Sci 2005;118:313–322.
Wong NA, Pignatelli M . Beta-catenin—a linchpin in colorectal carcinogenesis? Am J Pathol 2002;160:389–401.
Johnson V, Volikos E, Halford SE, et al. Exon 3 beta-catenin mutations are specifically associated with colorectal carcinomas in hereditary non-polyposis colorectal cancer syndrome. Gut 2005;54:264–267.
Acknowledgements
This study was supported by a grant from the Swiss National Foundation (Grant no. PBBSB-110417) and the Novartis Foundation, formerly Ciba-Geigy-Jubilee-Foundation. We thank Privatdozent Dr Hanspeter Spichtin, Institute of Clinical Pathology Basel, Switzerland and Professor Dr Robert Maurer, Institute of Pathology, Stadtspital Triemli, Zuerich, Switzerland for providing the cases.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lugli, A., Zlobec, I., Günthert, U. et al. Overexpression of the receptor for hyaluronic acid mediated motility is an independent adverse prognostic factor in colorectal cancer. Mod Pathol 19, 1302–1309 (2006). https://doi.org/10.1038/modpathol.3800648
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800648