Abstract
The p16INK4a is a cyclin-dependent kinase inhibitor that decelerates the cell cycle by inactivating the cyclin-dependent kinases involved in the phosphorylation of the retinoblastoma protein (RB). Expression of E6 and E7 oncogenes of high-risk (HR) human papillomavirus (HPV), affecting the RB-p16 pathway, leads to p16 upregulation. Although it is widely reported that p16 is overexpressed in a high percentage of preneoplastic lesions and in almost all carcinomas of the uterine cervix, protein upregulation and its correlation with HPV infection in low-grade lesions is still being debated. In this study, we investigated in parallel, p16 expression and HPV infection in 100 cervical biopsies (17 normal tissues, 54 CIN1, 10 CIN2, 11 CIN3, eight invasive squamous cancers). Results obtained demonstrated that none of the 17 normal cervical tissues, evaluated by immunohistochemistry, presented p16 positivity whereas, starting from CIN1 (31%) to CIN2 (90%), CIN3 (100%) and carcinomas (100%), a constant and significant increase of protein overexpression (P<0.0001) was observed. In addition, p16 overexpression consistently showed elevated sensitivity (84%) and specificity (98%) in detecting HR-HPV infection with a high positive predictive value (97%) and negative predictive value (86%). Of interest, 93% of the p16-positive CIN1 were also HR-HPV infected. Our findings confirmed that p16 overexpression is associated to high-grade precancerous lesions and cervical carcinomas, and further demonstrated that immunohistochemical evaluation of p16 may be a useful biomarker in identifying HR-HPV-infected low-grade lesions.
Similar content being viewed by others
Main
Cancer of the uterine cervix is one of the most common cancers among women worldwide and second only to breast cancer in incidence and mortality.1 Epidemiological and molecular studies have shown that human papillomavirus (HPV) infection is the most important factor for cervical carcinogenesis. Recent extensive studies have revealed the existence of more than 100 different HPV genotypes of which about 30 can infect the uterine cervical epithelium.2 Each HPV subtype has been shown to have a different carcinogenetic potential on cervical epithelial cells and has been subdivided into high- (HR) and low-risk (LR) types. Although the persistence of viral infection and the capability of viral DNA integration into the host cellular DNA are closely associated with malignant transformation,3, 4, 5 the reasons for the difference in the oncogenic potential of each HPV subtype remains unresolved. It has been suggested that the oncoproteins encoded by the E6 and E7 viral oncogenes, which bind important host regulatory proteins (ie p53 and retinoblastoma protein (RB)), play a pivotal role in cervical carcinogenesis. In fact, p53 protein is degraded following E6 binding, whereas hypophosphorylated RB is functionally inactivated and degraded by E7 binding. In particular the E7 gene product from various HPV types has different efficiency both in RB binding and degradation.6, 7 The E7-RB binding favours the release of E2F-like transcription factors over the active RB protein. As a consequence, the transcription of RB-regulated genes is activated, promoting the G1/S-phase progression of the cell cycle. Thus, E7 mimics the inactivation of RB due to the phosphorylation induced by the cyclin-dependent kinases CDK4 and CDK6. In normal cells, CDK4 and CDK6 activity is regulated by several CDK inhibitors including p16. E2F-1 accumulation, a consequence of RB phosphorylation, leads to p16 upregulation which allows the control of the kinase activity through a feedback inhibition mechanism.7 Since p16 expression is negatively controlled by an active RB, reduction or loss of the hypophosphorylated protein can induce p16 overexpression. Consequently, HPV E7 RB inactivation may result in the enhanced expression of p16 which appears to be a marker of E7 gene activity. Therefore the immunohistochemical (IHC) evaluation of p16 protein, known to be widely detected in cervical carcinomas, could improve conventional histopathological diagnosis of preneoplastic diseases,7, 8, 9, 10, 11, 12, 13, 14, 15, 16 thus increasing interobserver agreement in the identification of low-grade lesions.11 However, only a few studies have focused their attention on the potential role of this protein also in identifying those low-grade lesions caused by HR-HPV infection, which could be at higher risk of transformation.9, 13, 17 The present study aimed at evaluating p16 overexpression in parallel with HPV infection, in a series of 100 cervical biopsies comprising normal tissues, preneoplastic and neoplastic lesions. The potential association between p16 and HPV infection was investigated to verify whether p16 upregulation could be a valuable tool in the identification of dysplastic lesions caused by HR-HPV genotypes.
Materials and methods
Patients
We analyzed 100 formalin-fixed cervical biopsies obtained from the Regina Elena Cancer Institute archives between 2000 and 2003: 17 histological normal tissues, 54 CIN1, 10 CIN2, 11 CIN3 and eight invasive squamous carcinomas. Patient median age was 41 years (range 18–82 years). All hematoxylin–eosin slides were re-examined independently by two investigators without any pathological or clinical information concerning the cases under study.
Cell Line Preparation
CaSki, HeLa, C33A and A431 cell lines were kindly provided by the Virology Laboratory of the Regina Elena Cancer Institute (Rome, Italy). CaSki and HeLa are both cervical carcinoma-derived cell lines containing HPV 16 DNA (500–600 copies per cell) and HPV 18 DNA (10–50 copies per cell), respectively. C33A and A431 are HPV-negative cancer cell lines derived from cervical and cutaneous squamous carcinomas, respectively. These four cell lines were grown to confluence and cells were harvested by treatment with trypsin-EDTA solution. After washing in phosphate buffer solution 10 mM pH 7.4 (PBS), cells were pelleted and fixed overnight in 4% buffered formalin. The cells were then dehydrated, cleared and embedded in paraffin according to conventional histological techniques.
p16 Immunostaining
The p16 immunostaining was performed on 5 μm sections from paraffin blocks by means of a commercially available kit (DakoCytomation, Milan, Italy), including the Monoclonal Antibody (MoAb) E6H4 which identifies an epitope between aa 134 and 156 of the p16 protein.8 A specific epitope retrieval method was carried out by heating the slides immersed in the kit retrieval solution in a calibrated water bath at 96°C for 40 min. Nuclear as well as cytoplasmic reactivity was considered positive. Staining intensity was not graded to avoid subjective interpretation. p16 did not show any reaction with normal epithelial or mesenchymal cells and only sporadic staining of endocervical metaplasic cells was evidenced (Figure 1).
HPV Testing
The presence and typing of HPV DNA was assayed by polymerase chain reaction (PCR) and subsequent reverse dot-blot hybridization with sequence-specific oligonucleotide probes (Diatech, Iesi, Italy). DNA was isolated from 20 μm paraffin sections, using a commercial extraction kit (Helix Fast DNA kit; Diatech) according to the manufacturer's specified protocol. Briefly, after dewaxing, the desiccate sections were incubated in digestion solution containing proteinase K, at 56°C overnight. Proteinase K was then inactivated heating at 94°C for 10 min. The extracted DNA was stored at −20°C till use. For the amplification, we used a general HPV degenerate primer set that detects sequences within the L1 open reading frame of at least 23 HPV types, including the 18 genotypes at present considered HR (HPV 16, 18, 31, 33, 34, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, 82) and five LR types (HPV 6, 11, 40, 43 and 44). The 145 bp amplified gene fragment is characterized by hybridization reaction with sequence-specific oligonucleotides representing particular HPV strains. The amplified DNA is denaturated and brought into contact with nitrocellulose strips where gene probes for different HPV strains, as well as control zones, are lined. During hybridization the denaturated DNA binds to the gene probes on the strips. The gene probe biotin-marked amplicon hybrids can then be detected with streptavidin-coupled alkaline phosphatase. A positive reaction can be seen as dark staining in the reaction zone. Each strip has a conjugate- and an amplification-control zone that must be completely developed during the test. The amplification-control zone contains a probe for the GAP-DH housekeeping gene which is present in each human genomic DNA sample, and is a control for correct DNA extraction and amplification. A HPV-poly zone is developed if there are any of the 23 HPV genotypes, described above, present in the DNA sample, and an HR-HPV zone is developed if there is one or more HR types. Specific single bands characterize the genotypes 16, 18 and 45. A single reaction zone is developed when one or more of the 31, 33, 34, 35 and/or 39 genotypes is present, and another single reaction zone is developed when one or more of the 51, 52, 53, 56, 58 and/or 59 genotypes is present. When only the HPV-poly and HPV-HR zones are developed, sequences of one or more of the 66, 68, 73 and/or 82 genotypes are present in the sample. The HPV-poly- and LR-specific bands are developed together when one or more of the five LR types, detected by the kit, is present.
Statistical Analyses
The p16 immunoreactivity was evaluated in terms of sensitivity, specificity and predictive value of the test for either detection of CIN or a worse lesion, or for HR-HPV infection, according to Galen and Gambino.18 A Fisher's exact test was applied to assess associations between variables and these were considered significant whenever the two-sided test had a <0.05 P-value. The Kappa statistic was used to assess the degree of agreement between p16 and HPV tests. Kappa values >0.75 are regarded as excellent agreement beyond chance, values <0.40 as poor agreement beyond chance, and values between 0.40 and 0.75 as fair to good agreement beyond chance.19
Results
HPV Detection and p16 Immunostaining on Human Cancer Cell Lines
In order to evaluate the sensitivity and specificity of the technical procedures used in this study, HPV testing and p16 immunohistochemistry were first assayed in four different cancer cell lines (see Materials and methods, Table 1). As expected, two cell lines, C33A and A431, were HPV negative, whereas the other two cell lines, CaSki and HeLa, harbored 16 and 18 HPV DNA sequences, respectively. None of the four cell lines showed any reactivity with the LR-HPV probe. In contrast to CaSki and HeLa cells, which were homogeneously p16 positive, A431 cells were consistently p16 negative. However, the HPV-negative C33A cells, as already reported by other authors,12, 13 unexpectedly displayed p16 positivity.
p16 Distribution
Figure 2 shows p16 distribution in the 100 cervical biopsies. None of the 17 normal cervical tissues, evaluated by IHC, demonstrated p16 positivity whereas, starting from CIN1 through to carcinomas, a constant and significant increase of protein overexpression was observed (P<0.0001). CIN1, as well as the CIN2 lesions were in fact positive in 17 out of 54 (31%) and nine out of 10 (90%) of the cases, respectively, while both the CIN3 lesions and the squamous carcinomas displayed a strong and diffuse nuclear and/or cytoplasmic staining for p16 in all the analyzed cases. Furthermore, p16 overexpression demonstrated a 54% sensitivity with a 100% specificity in detecting CIN1-3 lesions as well as cervical carcinomas. Therefore, p16 positive predictive value (PPV) was particularly high (100%), while a low negative predictive value (NPV) was detected (31%).
HPV Distribution
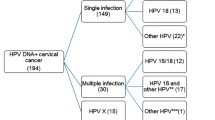
A total of 86 out of the 100 cases included in our study were analyzed by PCR for HPV infection (15 normal tissues, 48 CIN1, 10 CIN2, five CIN3 and eight squamous carcinomas). Of the 86 samples tested, 43 had HR-HPV sequences (50%) and three cases LR-HPV sequences (3.5%). As described in Figure 3a, in normal tissues only one out of 15 (7%) showed HR-HPV infection. In the CIN1, HR-HPV genotypes were found in 19 of the 48 lesions tested (39.5%), whereas three lesions were infected by LR types (6.2%). All the CIN2 and CIN3 lesions and the squamous carcinomas analyzed displayed HR genotypes. The increase of HR-HPV presence during progression from mild, intermediate and severe dysplasia to invasive disease was statistically significant (P<0.0001).
When we analyzed the distribution of the different HR-HPV genotypes in our cohort of patients (Figure 3b), we found that the 16 genotype was prevalent (48%) followed by the 50s (18%), 30s (9%) and 18 (7%) genotypes. In four cases (9%) we found one or more of the 66, 68, 73 and 82 genotypes. Finally, a coinfection was found in four samples (9%). In detail, two cases displayed HR/LR type coinfection and two HR/HR coinfection.
Relationship between p16 Immunostaining and HPV Distribution
In order to verify whether a relationship between p16 overexpression and HR-HPV infection occurs in our series of cervical samples, we compared the IHC findings with the presence of HR-HPV. Figure 4 (panel a/b) summarizes the results obtained. None of the 15 morphologically normal tissues were p16 positive though one specimen was HR-HPV positive. In all, 15 out of 48 CIN1 tested were p16 positive (31%), of which 93% (14 out of 15) were HR-HPV infected. Five CIN1 cases were p16 negative and HR-HPV positive. The majority of the p16-positive high-grade lesions were HR-HPV infected (90% CIN2 and 100% CIN3 and carcinomas). The correlation between p16 upregulation and HR-HPV infection was statistically significant (K=0.81, P<0.0001). Therefore, p16 overexpression consistently evidenced high sensitivity (84%) and specificity (98%) in detecting HR-HPV infection with a high PPV (97%) and NPV (86%).
Discussion
The causal role of human HPV infection in cervical cancer has been documented beyond reasonable doubt and HPV has been proposed as the first ever identifiable necessary cause of a human cancer.1 Moreover, epidemiological studies have revealed that HR-HPV infections are very common, particularly in young women, but the prevalence of infection decreases after the age of 30 years, suggesting that in most women the infection is self-limited and spontaneously resolves without leading to relevant pathologies.3 On the other hand, there is a lot of confusion about molecular assays for HPV analysis that are not standardized and rarely subjected to quality control programs. Moreover, even when a lesion is present, there is a substantial lack of interobserver reproducibility in the histological diagnosis of CINs, and the lack of interpathologist agreement is most evident for less severe interpretations,11, 20, 21 particularly in distinguishing CIN1 from reactive lesions. Moreover, although cervical intraepithelial lesions conventionally are subjectively divided into three grades, the histological features represent a diagnostic continuum, one stage merging imperceptibly with the next.22 These considerations highlight the need to identify specific biomarkers with a significant positive predictive value in detecting the presence of cervical dysplasia or cancer.23 p16 could be a useful biomarker, capable of complementing conventional morphology. In fact, in cervical carcinogenesis, HR-HPV E7 activity leads to p16 overexpression which may be easily detected by immunohistochemistry and its presence is uniform across HR-HPV types.
In order to verify whether p16 is valuable as a biomarker in the identification of those HR-HPV-infected lesions which could be at higher risk of developing cancer, in this study, we evaluated p16 overexpression in parallel with HPV infection in a series of 100 cervical biopsies including histological negative cases, low- and high-grade lesions, as well as squamous cervical carcinomas. Our data demonstrated that histologically negative tissues were always p16 negative and, in the majority of cases (93%), also HR-HPV negative, with a 86% NPV. Otherwise, 31% of the CIN1 lesions displayed p16 immunoreactivity and 93% of these p16-positive CIN1 were also infected by HR-HPV genotypes. The correlation between p16 overexpression and HR-HPV infection significantly and progressively increases in CIN2 (90%), CIN3 and carcinomas (100%). In our study almost all the HPV-negative lesions were also p16 negative. In fact, only one case, morphologically diagnosed as CIN1, was p16 positive and HR-HPV negative.
However, we also found HR-HPV infection in seven biopsies which were p16 negative (one normal tissue, five CIN1 and one CIN2). There are a number of considerations to explain these findings. In fact, as widely reported in other solid tumors,24, 25, 26 p16 gene mutation, deletion or hypermetylation of the promoter, could avoid p16 expression, although, in cervical carcinogenesis, these alterations are rare events. In addition, HPV, although identified by the test, could be inactivated by the host immunosurveillance, the infection could be very recent or the virus could be defective. In all these cases, the transcription of the E6 and E7 oncogenes fails and the RB-p16 pathway is not affected hampering p16 overexpression. These limitations could be overcome by analyzing the expression of the HR-HPV oncogenes E6 and E7 that are required to induce and maintain neoplastic growth in cervical epithelium.4, 27, 28 However, attempts to monitor viral oncogene expression, either at the RNA or protein levels, in routine clinical samples, have been hampered by the relatively low expression level of the viral oncogene products, the lack of sufficiently specific technologies or the availability of reagents sensitive enough to detect E6 and E7.11 Moreover, IHC detection of the E6 and E7 gene products may be difficult due to the very short half-life and high turnover rates of these proteins which are degraded within several minutes after translation.23
However, the strong p16 immunostaining we observed in almost all the cervical high-grade lesions and cancers indicated that p16 gene inactivation is a rare event in cervical carcinogenesis supporting the hypothesis that p16 upregulation is a consequence of HR-HPV infection. Although our data is generally in agreement with that obtained in high-grade lesions and cancer by other authors, a higher percentage of p16-positive CIN1 than we found has been reported.7, 8, 9, 10, 11, 12, 13, 14, 15, 16 This may be due to methodological differences, such as the use of different antibodies for immunohistochemistry, which sometimes makes it difficult to compare results. As regards HPV assays, the virus was detected on histological samples by means of in situ hybridization or PCR. Also in the latter case different primers set and different technical approaches were used. Likely due to the lack of methodological standardization, the prevalence of HR-HPV CIN1 is variable among studies ranging from 32 to 80%.9, 10 Our results fall in this range, but the number of HR-HPV-positive CIN1 cases is lower than those found by the majority of the studies. Possible explanations for these findings are mainly a probable overdiagnosis of low-grade lesions. Therefore, studies measuring interobserver variability of histological diagnosis of cervical lesions, evidenced that the most significant discrepancies are just in the ability of the pathologists to distinguish CIN1 from reactive squamous proliferation.21
In this context, p16 testing can substantially improve the conventional morphological diagnosis of cervical preneoplastic lesions.11 Nevertheless, only a few studies have analyzed the correlation between the p16 overexpression and HPV infection.7, 9, 10, 12, 13, 14, 15, 16, 17 It is noteworthy that most low-grade lesions spontaneously clear 6–12 months after appearance, probably due to immunological intervention.29 Morphological criteria alone are not sufficient to distinguish lesions that may regress from those that might persist and progress. In fact, as demonstrated in other solid tumors,30, 31 in cervical precursor lesion, independent of morphological features, molecular alterations, hastening tumor development, may occur. In this context, the evaluation of HR-HPV infection as well as p16 immunoreactivity, could be useful tools of particular clinical value in identifying cases with a higher tendency to progress to high-grade lesions. One hypothesis could be that the HR-HPV-negative low-grade lesions have a higher probability of disappearing with respect to HR-HPV-positive ones. Of interest, it was recently reported that CIN1 cases with diffuse p16 staining have a significantly greater tendency to progress to a high-grade lesion than p16-negative cases.32 Nevertheless, in this study no data about HPV infection is available. In our study, we show that a statistically significant inter-relationship occurs between p16 overexpression and HR-HPV infection (K=0.81) with an 84% sensitivity, 98% specificity, 97% PPV and 86% NPV. Particularly, our data demonstrated that almost all the p16-positive CIN1 lesions were also HR-HPV positive (93%). This is a finding of particular clinical interest. In fact, controversial data are provided about the correlation between p16 overexpression and HR-HPV infection in CIN1.7, 8, 10, 12, 15, 16
In particular Murphy recently showed that all the CIN1 lesions he tested were p16 positive even though only 55% were HR-HPV infected13 and Guimaraes, in a selected and limited cohort of patients, did not find any correlation between low-grade lesions and HR-HPV infection.17 On the other hand, our results are consistent with Kalof's conclusions, even if he tested a smaller cohort of patients focusing on ISH HPV assay.9 Our data, obtained on a larger number of cases, demonstrated that p16 may represent a specific and sensitive biomarker for cervical cells with active expression of HPV oncogenes and might play a pivotal role in identifying low-grade lesions strongly associated with the presence of HR-HPV infection. In our opinion p16-positive HR-HPV-infected CIN1 are the low-grade lesions which need particular clinical attention. Finally, this method, applied to cytology, might also prove to be useful in reinforcing HPV DNA testing for the management of patients with abnormal pap smears or in primary screening.
References
Bosch FX, Lorincz A, Munoz N, et al. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 2002;55:244–265.
Munoz N, Bosch FX, De Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003;348:518–527.
Daltein V, Riethmuller D, Pretet J-L, et al. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal french cohort study. Int J Cancer 2003;106:396–403.
Fehrmann F, Laimins LA . Human Papillomavirus: targeting differentiating epithelial cells for malignant transformation. Oncogene 2003;22:5201–5207.
Walboomers JMM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–19.
Giarrè M, Caldeira S, Malanchi I, et al. Induction of Rb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a-imposed G1 cell cycle arrest. J Virol 2001;75:4705–4712.
Keating JT, Cviko A, Riethdorf S, et al. Ki-67, Cyclin E, and p16INK4a are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol 2001;25:884–891.
Agoff N, Lin P, Morihara J, et al. p16INK4a expression correlates with degree of cervical neoplasia: a comparison with Ki-67 expression and detection of high risk HPV types. Mod Pathol 2003;16:665–673.
Kalof AN, Evans MF, Simmons-Arnold L, et al. p16INK4a immunoexpression and HPV in situ hybridation signal patterns. Am J Surg Pathol 2005;29:674–679.
Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16INK4a as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 2001;92:276–284.
Klaes R, Benner A, Friedrich T, et al. Immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol 2002;26:1389–1399.
Murphy N, Ring M, Killalea AG, et al. p16INK4a as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep™ smears. J Clin Pathol 2003;56:56–63.
Murphy N, Ring M, Heffron CCBB, et al. p16INK4a CDC6 and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J Clin Pathol 2005;58:525–534.
Sano T, Oyama T, Kashiwabara K, et al. Immunohistochemical overexpression of p16 protein associated with intact retinoblastoma protein expression in cervical cancer and cervical intraepithelial neoplasia. Pathol Int 1998;48:580–585.
Sano T, Oyama T, Kashiwabara K, et al. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol 1998;153:1741–1748.
Sano T, Masuda N, Oyama T, et al. Overexpression of p16 and p14ARF is associated with human papillomavirus infection in cervical squamous cell carcinoma and dysplasia. Pathol Int 2002;52:375–383.
Guimaraes MCM, Goncalves MAG, Soares CP, et al. Immunohistochemical expression of p16INK4a and bcl-2 according to HPV type and to the progression of cervical squamous intraepithelial lesions. J Histochem Cytochem 2005;53:509–516.
Galen RS, Gambino SR . Beyond Normality: the Predictive Value and Efficiency of Medical Diagnoses. Wiley: New York, 1972.
Fleiss JL . Statistical Methods for Rates and Proportions. John Wiley & Sons: New York, 1981.
Stoler MH, Schiffman M . Atypical Squamous Cells of Undetermined Significance-Low grade Squamous Intraepithelial Lesion Triage Study (ALTS) Group. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. JAMA 2001;285:1500–1505.
Wright TC, Kurman RJ, Ferenczy A . Precancerous lesions of the cervix. In: Kurman RJ (ed). Blaustein's Pathology of the Female Genital Tract, 4th edn. Inc. Springer-Verlag: New York, 1994, pp 229–277.
Wells M, Ostor AG, Crum CP, et al. Epithelial tumors. In: Tavassoli FA, Devilee P (eds) World Health Organization Classification of Tumors, Tumors of the Breast and Female Genital Organs. IARC Press: Lyon, 2003, pp 262–279.
von Knebel Doeberitz M . New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer 2002;38:2229–2242.
Esposito V, Baldi A, Tonini G, et al. Analysis of cell cycle regulator proteins in non-small cell lung cancer. J Clin Pathol 2004;57:58–63.
Ghiorzo P, Villaggio B, Sementa AR, et al. Expression and localization of mutant p16 proteins in melanocytic lesions from familial melanoma patients. Hum Pathol 2004;35:25–33.
Kawaguchi K, Oda Y, Saito T, et al. Mechanisms of inactivation of the p16INK4a gene in leiomyosarcoma of soft tissue: decreased p16 expression correlates with promoter metylation and poor prognosis. J Pathol 2003;201:487–495.
von Knebel Doeberitz M, Oltersdorf T, Schwarz E, et al. Correlation of modified human papilloma virus early gene expression with altered growth properties in C4-1 cervical carcinoma cells. Cancer Res 1988;48:3780–3786.
zur Hausen H . Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin Cancer Biol 1999;9:405–411.
zur Hausen H . Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev 2002;2:342–350.
Xu R, Perle MA, Inghirami G, et al. Amplification of HER2/neu gene in HER2/neu-overexpressing and -nonexpressing breast carcinomas and their synchronous benign, premalignant and metastatic lesions detected by FISH in archival material. Mod Pathol 2002;15:116–124.
Mottolese M, Nadasi EA, Botti C, et al. Phenotypic changes of p53, HER2, and FAS system in multiple normal tissues surronding breast cancer. J Cell Physiol 2005;204:106–112.
Negri G, Vittadello F, Romano F, et al. p16INK4a overexpression and progression risk of low-grade intraepithelial neoplasia of the cervix uteri. Virchows Arch 2004;445:616–620.
Acknowledgements
This study has been supported by Translational Research Group in Gynecological Oncology of the Regina Elena Cancer Institute, and by Lega Italiana per la Lotta contro i Tumori.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Benevolo, M., Mottolese, M., Marandino, F. et al. Immunohistochemical expression of p16INK4a is predictive of HR-HPV infection in cervical low-grade lesions. Mod Pathol 19, 384–391 (2006). https://doi.org/10.1038/modpathol.3800551
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800551
Keywords
This article is cited by
-
Efficacy and safety of an adsorbent and anti-oxidative vaginal gel on CIN1 and 2, on high-risk HPV, and on p16/Ki-67: a randomized controlled trial
Archives of Gynecology and Obstetrics (2021)
-
p16 staining has limited value in predicting the outcome of histological low-grade squamous intraepithelial lesions of the cervix
Modern Pathology (2016)
-
Expression of P16 in high-risk human papillomavirus related lesions of the uterine cervix in a government hospital, Malaysia
Diagnostic Pathology (2014)
-
Claspin as a biomarker of human papillomavirus-related high grade lesions of uterine cervix
Journal of Translational Medicine (2012)
-
p16 expression is not associated with human papillomavirus in urinary bladder squamous cell carcinoma
Modern Pathology (2012)