Abstract
The protein encoded by the MDM2 oncogene inhibits the function of p53, leading to increased cell growth, avoidance of apoptosis, tolerance of genetic instability, and resistance to chemotherapy. The present study was performed to evaluate the relationship between MDM2 protein expression and survival in breast carcinoma. Two series of cases were used in this study: the first to identify the cutoff to be used in the interpretation of MDM2 immunostaining and perform preliminary survival analysis, and a second, independent series, to validate the findings from the first series and to perform multivariate analysis. For both series, archival sections of tissue microarrays were stained with anti-MDM2 antibody (NeoMarkers, Fremont, CA, USA) and MDM2 staining intensity was scored semiquantitatively. In the first series, 49 of 362 (14%) interpretable cases were positive for MDM2 expression, with 35 (10%) showing weak positivity and 14 (4%) strong positivity. Patients with MDM2-positive tumours had a significantly worse disease-specific survival than patients with MDM2-negative tumours (P=0.0022, 10-year DSS 61% (95% CI: 45–73) vs 73% (95% CI: 67–77)). No significant difference in survival was observed between patients with strongly and weakly MDM2-positive tumours (P=0.3). Accordingly, in the independent validation series weak and strong MDM2 positivity were combined and considered to be MDM2 positive. MDM2 expression was seen in 230/1747 (13%) interpretable cases in this series, with a significant difference (P<0.0001) in DSS between MDM2-negative and MDM2-positive cases (10 year DSS 58% (95% CI: 51–64) vs 73% (95% CI: 70–75)). MDM2 was an independent prognostic marker (HR=1.35, P=0.02) in a Cox regression model including MDM2 expression, tumour grade, nodal status, ER status and tumour size. Immunohistochemical studies of MDM2 in more than 2000 breast carcinomas show that MDM2 is an independent negative prognostic marker.
Similar content being viewed by others
Main
The human MDM2 gene encodes a 491 amino-acid protein that contains a binding domain for the tumour suppressor p53.1 This 90 kDa nuclear phosphoprotein can form a complex with both mutant and wild-type p53,2 and takes part in downregulation of p53 functions, promoting the rapid degradation of p53 via a ubiquitin–proteasome pathway.3, 4, 5 MDM2 protein acts as a nuclear-cytoplasmic transporter for the p53 protein.5 The MDM2 gene is transcriptionally upregulated by p535, 6, 7 and acts as an oncogene in tissue culture.5 In human tumours, MDM2 takes part in tumorigenesis through one of the three possible mechanisms: gene amplification, increased transcription or enhanced translation.5
Overexpression of MDM2 is described in many human cancers, including breast carcinoma.1, 3, 8 Overexpression of MDM2 protein correlates with high grade and was found to be an independent negative prognostic marker in human breast cancer in one study.9 Other investigators observed that MDM2 overexpression correlates with favourable prognostic parameters such as ER overexpression.10 Studies of MDM2 protein expression in breast carcinoma and its prognostic significance have been based on a limited number of cases, and the results of these studies have been inconsistent, as mentioned above. MDM2 amplification in breast carcinoma was seen in 6% of cases, in a study of greater than 2000 cases.11 Amplification of the MDM2 gene did not correlate with disease-specific survival (P=0.26) in this study. However, gene amplification is only one of the mechanisms of MDM2 overexpression, and definitive studies on the significance of MDM2 protein expression in breast carcinoma have not been performed. Thus, MDM2 protein has a number of functions important in breast cancer progression but translational studies on the relationship between MDM2 expression and survival of breast carcinoma patients are necessary to determine its prognostic significance and correlations with other oncoproteins.
Promising results in initial studies of novel biomarkers frequently prove to be irreproducible. For a biomarker to be clinically relevant, it has been proposed that there should routinely be independent studies of an initial cohort of patients and then a second cohort; the latter must be analyzed as a stand-alone validation set that is not influenced by the results obtained in the original cohort.12 The use of standardized, reproducible assays with predefined scoring systems and cut points is particularly important.12
In the present study, immunohistochemical expression of MDM2 was evaluated as a prognostic marker and its relationship with other clinical and pathological parameters determined. Two separate series of cases were used, totalling more than 2000 cases with interpretable MDM2 immunostaining results, to allow definitive analysis of the significance of MDM2 protein overexpression in breast carcinoma.
Materials and methods
Case Selection
Cases used in the analysis were women who were diagnosed with invasive breast carcinoma. Two groups of cases were included into the study: (1) an initial series of 438 women with primary invasive breast cancer who underwent surgery for breast cancer between 1974 and 1995 at Vancouver General Hospital, and (2) a second, independent series of 2651 cases with the same diagnosis collected from the British Columbia Cancer Agency from 1989 to 1992. Outcome data were available for all cases. Ethical approval to perform this study was granted by the ethical review board of British Columbia Cancer Agency and the University of British Columbia. Although there were 438 patients in the initial series, and 2651 patients in the validation series, the interpretable number of cores was less than these numbers, depending on the tissue microarray slide. Causes of missing cores included tissue falling off the slides, insufficient or absent tumour tissue within the cores, or artefactual distortion making interpretation impossible. There were 362 interpretable cores in the initial series in the slides, stained for MDM2, and 1747 interpretable cores in the validation series. When performing pairwise correlations between the expression of MDM2 protein and other biomarkers, only the cores preserved in both stains were taken into account.
Tissue Microarrays and Immunostaining
Tissue microarrays (TMA) were built using tissue cores from formalin-fixed paraffin-embedded tumours. Hematoxylin and eosin (H&E)-stained slides were reviewed, and areas containing tumour tissue were marked on both the slides and corresponding paraffin blocks for tissue microarray construction. Duplicate 0.6 mm tissue cores per case were used for building the screening array, and single 0.6 mm cores per case were taken for the second validation array. Microarray blocks were constructed using a manual arrayer (Beecher Instruments, Inc., Silver Springs, MD, USA) as previously described.13, 14, 15 Sections (4 μm thick) were cut from the array blocks. Antigen retrieval was performed by steaming for 30 min in citrate buffer (pH 6.0). Hybridization with the primary MDM2 antibody (mouse monoclonal anti-MDM2 antibody, Clone MSP14, NeoMarkers, Fremont, CA, USA) was carried out in a sealed immunochamber overnight at 4°C at 1:200 dilution. Detection was performed with the labelled anti-mouse polymer (DAKO, Carpinteria, CA, USA) for 30 min in the sealed immunochamber, followed by the Nova Red chromogen (Vector Laboratories, Burlingame, CA, USA) for 10 min. Slides were counterstained with hematoxylin and mounted.
All the slides were scanned using Bacus Laboratories Inc. Slide Scanner (BLISS) (Bacus Laboratories, Inc., Lombard, IL, USA), based on an Olympus microscope with motorized stage and autofocus capabilities. Slides were scanned at × 20 objective magnification; nine images per core (3 × 3) were taken with pixel dimensions 752 × 480 each. Therefore, total pixel dimensions per core were 2256 × 1440. WebSlide Browser along with SlideTray programme (Bacus Laboratories, Inc, Lombard, IL, USA) was used for viewing preview images of the arrays and images of individual cores. Hematoxylin and eosin slides were scanned along with immunostained ones to ensure correspondence of the immunostained structures to the breast carcinoma. All the images of the slides are stored at our internet website and these are publicly available for viewing and scoring.16 The web application consists of two components: (1) GPEC image relational database that stores the cores information, and the expression levels; (2) the Web application that allows user to do queries and send request to the WebSlide server for displaying dynamic images by java applet. This website is accessible through https://www.gpecimage.ubc.ca/tma/web/viewer.php.
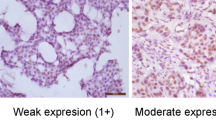
Scoring of MDM2 immunostaining was performed semiquantitatively, using digital images and 22-in monitor with hardware calibration capabilities. Staining was considered to be negative (0) if no staining was seen within the tumour, weak positive (1+) if focal staining was seen, and strong positive (2+) if there was a diffuse staining in more than 80% of tumour cells. Nuclear staining could be detected in very few cases, and the vast majority of positive cases showed only cytoplasmic staining; therefore, cytoplasmic staining only was taken into account when scoring. Scores were entered into a standardized Excel worksheet (Microsoft Excel, Microsoft, Redmond, WA, USA) with a sector map matching each TMA section.
Scores for the duplicate cores were consolidated to a single value per case using an Excel macro developed by DT. Where there were discrepant scores for the two cores, the higher value was accepted for the case. Cases were not included into statistical analysis if there was no interpretable data, that is, if there was no tumour tissue in the cores or the cores were cut through. Original scoring tables were deconvoluted using Deconvoluter 1.0717 and the resulting text files imported into SPSS 11.0 for Windows.
Statistical Analysis
Statistical analysis was performed in SPSS 11.0 for Windows (SPSS Inc., Chicago, IL, USA). Univariate analysis of survival was carried out by using Kaplan–Meier survival curves and log-rank statistics to test for significant differences between curves. Multivariate tests were performed using a proportional hazards (ie Cox regression) model. We used two-tailed Spearman's correlation to assess the relationship between the expression of MDM2 and other markers. All tests were two-sided and used a 5% alpha level to determine significance.
Results
The immunostaining pattern was predominantly cytoplasmic with very few cases showing nuclear staining. Weak to strong MDM2 expression was seen in the cytoplasm of the breast carcinoma cells (14% positive cases in the first array and 13% positive cases in the validation series). Representative photomicrographs of MDM2 immunostaining are shown as Figure 1. All images can be viewed at https://www.gpecimage.ubc.ca/tma/web/viewer.php.
Analysis of Data from the Initial Screening Set of 438 Patients
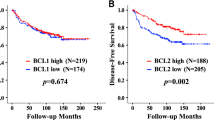
Survival analysis based on the initial set of breast carcinoma patients showed a significant difference when comparisons were pooled over strata (tumours with MDM2 negative, weakly positive and strongly positive expression, P=0.0045, Figure 2a). When comparing the groups pairwise, there was no significant difference between the survival of the patients with weak (n=35) and strong (n=14) expression of the protein (P=0.29). Therefore, all the positive cases were then combined and compared to the tumours that were negative for MDM2. The disease-specific survival of these two groups of patients was significantly different (P=0.0022, Figure 2b and Table 1).
Disease-specific survival (DSS) for the patients from the first series of cases. (a) There is significant difference in DSS between patients with MDM2 negative, weakly positive, and strongly positive tumours (P=0.0045). Log rank test performed with comparisons pooled over strata, with no individual curves comparisons. There is however no significant difference between MDM2 weakly positive and strongly positive cases (P=0.29) in pairwise comparison. (b) Combining weakly and strongly MDM2-positive cases, there is a significant difference in DSS for patients with MDM2-negative vs -positive tumours (P=0.0022).
MDM2 expression showed a significant positive correlation with p53 immunostaining and grade, and a negative correlation with ER (Table 2). A positive correlation was also seen between MDM2 expression and expression of the proliferation marker Ki67, the basal subtype marker cytokeratin 5/6, and cyclin E.
Analysis of Data from the Validation Set of 2651 Patients
Survival analysis was performed on the data obtained from an independent validation set of 2651 patients. Based on the cutoff value for interpretation of MDM2 immunostaining determined in our study of initial series we did not distinguish between weak and strong expression of MDM2; these cases were collectively considered to be MDM2 positive. The disease-specific survival of the patients with MDM2-negative and -positive tumours was significantly different (P<0.0001, Figure 3, and Table 1).
A proportional hazards (ie Cox regression) model was fitted with the following variables: Nottingham grade 1 and 2 vs Nottingham grade 3, Nottingham grade 1 vs 2 and 3, tumour size >2 cm vs ≤2 cm (Table 3). Results indicated significance of MDM2 expression as an independent prognostic factor in this series (HR=1.35, P=0.02, Table 3). Lymph node status (HR=2.5, P<0.000001), tumour grade (HR=0.6, P=0.000003), and ER status (HR=0.74, P=0.004) were more significant prognosticators than MDM2 expression, while tumour size did not show statistical significance in the multivariate model (HR=1.0, P=0.24).
Discussion
MDM2 overexpression has been reported to be a negative prognostic factor for a number of human tumours, including invasive breast carcinoma. We studied cytoplasmic expression of the MDM2 protein in breast carcinoma cells in two independent series of cases. Defining a cutoff between ‘significant overexpression’ and ‘no significant overexpression’ by immunostaining is difficult, as seen in the many years required to develop reproducible and clinically relevant assays for estrogen receptor and Her-2. We used patient outcome as a basis for determination of the cutoff, combining cases with weak and strong MDM2 immunoreactivity as they were associated with similar patient outcomes. Having determined a cutoff value for a particular assay using one series of cases, it is important to validate that cutoff in an independent series of cases, to avoid the problem of ‘over-fitting’ the data in the initial series, where the strategy of determining an assay cutoff based on patient outcomes ensures that you will see a maximal effect of the assay as a prognosticator. Many biomarkers have been shown to be significant in an initial study but follow-up studies have not confirmed these initial promising results, and this is one possible explanation.18
Another potential pitfall in assessment of novel biomarkers is to do only univariate analysis in an initial series of cases. When multivariate analysis is subsequently performed the novel factor is shown to covary with a stronger, routinely assessed variable such as lymph node status, and not be of independent prognostic significance. Our results show that MDM2 can be a strong prognostic marker in the breast carcinoma patients independent of clinical and pathological factors of known prognostic significance, including Nottingham grade, tumour size, lymph node status, and ER status.
A final consideration in assessment of novel biomarkers is to test a sufficient number of cases to have statistical power to be able to detect significant differences. Our validation series has more than 95% power to detect a two-fold relative risk if a variable (eg MDM2 immunostaining) is present in more than 10% of the population. The development of tissue microarray technology has made it technically and financially feasible to perform studies of this scale, and should dramatically hasten development of new biomarkers. There is now ample evidence that for most, but not all, biomarkers, the amount of tumour in small tissue microarray cores gives results highly representative of those obtained using whole sections.19
Additional levels of validation for new biomarkers would include technical (ie other laboratories are able to reproduce the staining results) as well as showing the results are generalizable to patients from other geographic regions. There is now increasing interest in the use of multiple biomarkers, either through immunohistochemical detection of protein, or RNA-based technologies, to refine prognostication in individual patients.20, 21, 22 This represents a change from the traditional approach of using single biomarkers and appears to be an inevitable next step in risk assessment and treatment planning for newly diagnosed patients with breast cancer. We currently treat many patients with breast cancer who derive no benefit from their treatment. The ability to identify a subset of node-negative breast carcinomas that have a sufficiently low risk of recurrence that the morbidity and potential mortality associated with treatment are not warranted is just one illustration of how this approach could be used to improve patient management. MDM2 is clearly a strong candidate to be included in a panel of biomarkers based on the results presented herein.
MDM2 has been previously studied on whole sections of breast carcinomas in series of up to approximately 100 cases, and the finding of a negative prognostic effect in the current study is similar to what has been seen in some but not all previous studies.9, 10 Some of the previous studies were of insufficient size to reliably detect outcome differences associated with MDM2 positivity, as noted previously.
Amplification of MDM2 in breast carcinoma, as assessed by FISH, is not associated with a worse prognosis,11 indicating that gene amplification is not the mechanism underlying MDM2 protein overexpression observed by immunohistochemistry in this study. A single nucleotide polymorphism (SNP309) in the MDM2 promoter has been reported to lead to increased MDM2 protein through enhanced binding of the transcriptional activator SP1 to the promoter.21 We tested 15 of the MDM2-positive tumours in this series for the presence of SNP309 and found only 3/15 heterozygous for this SNP with no homozygotes, a frequency lower than previously reported21 in a group of healthy volunteers (A De Luca, unpublished data). This suggests that this SNP does not account for the MDM2 overexpression observed. There is, however, a strong positive correlation with p53 immunostaining (Table 2). High levels of p53 expression in turn correlate with mutations in the p53 gene and accumulation of mutant protein.8 It is possible that MDM2 expression in breast carcinoma is not a reflection of a genetic abnormality at the MDM2 locus but instead represents a host response to p53 loss of function; this is similar to the observation in cervical carcinoma that inactivation of Rb and p53 by HPV oncoproteins results in a dramatic increase in production of immunoreactive p16 by the tumour cells.23, 24 If this is the case, MDM2 immunostaining could be a valuable marker of loss of normal p53 function, and used in addition to direct assessment of the p53 gene and/or protein to identify abnormality in this critically important pathway.
In conclusion, the results of the present study confirm the value of MDM2 protein expression as an independent negative prognostic marker in breast carcinoma patients.
References
Saji S, Okumura N, Eguchi H, et al. MDM2 enhances the function of estrogen receptor α in human breast cancer cells. Biochem Biophys Res Comm 2001;281:259–265.
Oliner JD, Pietenpol JA, Thiagalingam S, et al. Oncoprotein MDM2 conceals the activation domain of suppressor p53. Nature 1993;362:857–860.
Kubbutat MH, Jones SN, Vousden KH . Regulation of p53 stability by Mdm2. Nature 1997;387:299–303.
Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296–299.
Freedman DA, Wu L, Levine AJ . Functions of the MDM2 oncoprotein. Cell Mol Life Sci 1999;55:96–107.
Momand J, Zambetti GP, Olson DC, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992;69:1237–1245.
Barak Y, Juven T, Haffner R, et al. mdm2 expression is induced by wild type p53 activity. EMBO J 1993;12:461–468.
McCann AH, Kirley A, Carney DN, et al. Amplification of the MDM2 gene in human breast cancer and its association with MDM2 and p53 protein status. Br J Cancer 1995;71:981–985.
Jiang M, Shao Z-M, Wu J, et al. p21/waf1/cip1 and mdm-2 expression in breast carcinoma patients as related to prognosis. Int J Cancer 1997;74:529–534.
Hori M, Shimazaki J, Inagawa S, et al. Overexpression of MDM2 oncoprotein correlates with possession of estrogen receptor alpha and lack of MDM2 mRNA splice variants in human breast cancer. Breast Cancer Res Treat 2002;71:77–83.
Al-Kuraya K, Schraml P, Torhorst J, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res 2004;64:8534–8540.
Clark GM . Interpreting and integrating risk factors for patients with primary breast cancer. J Natl Cancer Inst Monogra 2001;30:17–21.
Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:767–768.
Makretsov N, Gilks CB, Coldman AJ, et al. Tissue microarray analysis of neuroendocrine differentiation and its prognostic significance in breast cancer. Hum Pathol 2003;34:1001–1008.
Makretsov NA, Huntsman DG, Nielsen TO, et al. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin Cancer Res 2004;18:6143–6151.
Ng TL, Gown AM, Barry TS, et al. Nuclear beta-catenin in mesenchymal tumors. Mod Pathol 2005;18:68–74.
Liu CL, Prapong W, Natkunam Y, et al. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol 2002;161:1557–1565.
Allred DC, Harvey JM, Berardo M, et al. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol 1998;11:155–168.
Torhorst J, Bucher C, Kononen J, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 2001;159:2249–2256.
Paik S, Shak S, Tang GT, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Eng J Med 2004;351:2817–2826.
Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 2004;119:591–602.
Callagy G, Cattaneo E, Daigo Y, et al. Molecular classification of breast carcinomas using tissue microarrays. Diagn Mol Pathol 2003;1:27–34.
Keating JT, Cviko A, Riethdorf S, et al. Ki-67, cyclin E, and p16INK4 are complementary surrogate biomarkers for human papillomavirus-related cervical neoplasia. Am J Surg Pathol 2001;25:884–891.
Klaes R, Friedrich T, Spitko D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 2001;92:276–284.
Acknowledgements
CDB, TON and DGH were supported as Scholars through the Michael Smith Foundation For Health Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Turbin, D., Cheang, M., Bajdik, C. et al. MDM2 protein expression is a negative prognostic marker in breast carcinoma. Mod Pathol 19, 69–74 (2006). https://doi.org/10.1038/modpathol.3800484
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800484
Keywords
This article is cited by
-
FKBP12 is a predictive biomarker for efficacy of anthracycline-based chemotherapy in breast cancer
Cancer Chemotherapy and Pharmacology (2019)
-
Gossypol has anti-cancer effects by dual-targeting MDM2 and VEGF in human breast cancer
Breast Cancer Research (2017)
-
MDM2 Contributes to High Glucose-Induced Glomerular Mesangial Cell Proliferation and Extracellular Matrix Accumulation via Notch1
Scientific Reports (2017)
-
Loss of Diacylglycerol Kinase-Ζ Inhibits Cell Proliferation and Survival in Human Gliomas
Molecular Neurobiology (2016)
-
Effects of MDM2 inhibitors on vascular endothelial growth factor-mediated tumor angiogenesis in human breast cancer
Angiogenesis (2014)