Abstract
In malignant melanomas, the loss of cell cycle control is thought to be due to a lack of retinoblastoma protein (pRb)-activity. Members of the previously described family of retinoblastoma-binding proteins (RBPs) are supposed to act as pRb-modulating factors. Based on RNA-fingerprinting of normal human melanocytes, we previously described a new family member with high sequence homology to the retinoblastoma-binding protein-2 (RBP-2), termed RBP2-Homolog 1 (RBP2-H1). Based on its UVB responsiveness, it was hypothesized that this gene may also play a role in melanocytic tumors. In the present study, we can confirm by real-time RT-PCR (six common melanocytic nevi, five advanced nodular melanomas and seven melanoma metastases) and immunohistochemistry (tissue microarrays: 52 melanocytic nevi, 60 melanomas, 60 metastases; and conventional sections: five common nevi, four advanced nodular melanomas, five melanoma metastases) that RBP2-H1 expression is progressively downregulated in advanced and metastatic melanomas in vivo with a certain intratumoral heterogeneity. Whereas benign melanocytic nevi are RBP2-H1 positive in about 70% of the cases, a lack of RBP2-H1 expression was found in 90% of the primary malignant melanomas and 70% of the melanoma metastases, respectively. Interestingly, a similar deficiency can be found in glioblastomas, but not epithelial cancers. In accordance to the in vivo data, established melanoma cell lines exhibit low but heterogeneous levels of RBP2-H1 expression. By co-immunoprecipitation, we provide the first evidence that a subfraction of total RBP2-H1 can bind to pRb, which makes this protein a true pRb-interacting factor. We conclude that loss of RBP2-H1 is a common finding in the progression of malignant melanomas. Since a direct interaction of RBP2-H1 and pRb seems possible, the loss of RBP2-H1 may possibly contribute to uncontrolled growth in malignant melanomas.
Similar content being viewed by others
Main
The loss of cell cycle control in malignant melanomas is thought to be due to a lack of retinoblastoma protein (pRb)-activity and not to a lack of its expression or mutation.1, 2, 3 To find new pRb-binding proteins (RBPs) functioning as potential pRb-modulating factors, Defeo-Jones and co-workers screened a human expression cDNA library with a recombinant Rb probe and identified two novel proteins termed RBP-1 and RBP-2 (alternatively ‘RBBP1, RBP1; RBBP2, RBP2’). Both proteins contain highly conserved pRb-binding motifs, which show a striking homology with viral oncogenic proteins such as E7, large T and E1A.4
More recently, based on RNA-fingerprinting, we have described a novel homolog of RBP-2, termed RBP2-homolog 1 (RBP2-H1, NCBI Genebank Accession No. AF087481).5 The corresponding transcript was detected due to its UVB responsiveness in normal nontransformed human melanocytes. It encodes a protein with a 54% amino-acid identity with RBP-2. Further computerized sequence analyses revealed various highly conserved motifs with possible functional implications regarding pRb-modulation. Among those, two DNA-binding zinc-finger (leukemia-associated protein, LAP) motifs, a rhombotin-2 (RBTN2, LMO2) binding domain and a domain possibly mediating a direct binding and interaction with pRb (non-T/E1A-pRb-binding domain) are most remarkable. However, a direct binding to or interaction with pRb has not been demonstrated previously.
Two more RBP2-homolog transcripts have been recently described that are differentially regulated in breast carcinomas, RBBP2H1a6 and PLU-1.7, 8 More detailed data on the expression of most RBPs in normal and neoplastic tissues are still lacking. While some RBPs, that is, PLU-1 and RBP-1, seem to play a role in breast cancer,9, 10 a possible impact of many other RBPs including RBP2-H1 on human cancer progression has not been investigated, yet.
In our previous work, we presented preliminary data suggesting a progressive deficiency of RBP2-H1 transcripts in melanomas.5 In this study, we intended to confirm this deficiency (i) on the RNA level using real-time RT-PCR and (ii) on protein level by immunohistochemistry and Western blotting with appropriate antibodies. Since RBP-2 is reported to bind and stabilize active, hypophosphorylated pRb,11, 12 we were further interested whether its closest relative, RBP2-H1, may in fact bind to pRb and fulfills the criteria of a true pRb-binding protein.
Materials and methods
Tissue Samples, Cell Lines and RNA Sources
Fresh, uncultured tumor material of six benign melanocytic nevi, five advanced nodular melanomas and seven melanoma metastases was collected at the Department of Dermatology, University of Regensburg, Germany. Samples of five glioblastomas were a kind gift from Juergen Schlegel, Department of Neuropathology, TU Munich, Germany. Written consent of the patients was obtained prior to surgery. Detailed tumor data of the melanocytic samples and the technical details of microdissection and RNA extraction have been recently published.13 Control RNAs from nonmelanocytic tissues were obtained from Ambion Inc. (Austin, TX, USA) and BD Biosciences, (Heidelberg, Germany). A breast cancer cell line (MCF-7; DSMZ, Braunschweig, Germany) and four melanoma cell lines (IGR-1, MEL-HO, COLO-849 from DSMZ, (Braunschweig, Germany); and A375-SM, kind gift from K Gehlsen, (SKCC, San Diego, CA, USA14) were taken into culture. A375-SM, COLO-894, MEL-HO and MCF-7 were cultured in RPMI 1640 (PAN Biotech, Aidenbach, Germany), IGR-1 in Dulbecco's MEM (Biochrom AG, Berlin, Germany). For RNA extraction, the RNeasyTM Mini Kit (Qiagen, Hilden, Germany) was used according to the manufacturer's recommendations. RNAs were subjected to quality controls using the Agilent 2100 bioanalyzer (Agilent biotechnologies, Palo Alto, CA, USA).
Gene Expression Analysis
RT-PCR was performed using the Reverse Transcription KitTM (Promega, Madison, WI, USA) according to the recommended protocol. For relative quantitation of transcribed gene specific RNA, TaqManTM real-time RT-PCR (Applied Biosystems, Darmstadt, Germany) was performed as described.15 Primers and probes for RBP2-H1 and RBBP2H1a were custom designed by Applied Biosystems (Assay by designTM). RBBP2H1a was analyzed to get information whether RBP2-H1 is specifically regulated in melanoma among the possible splice variants.
Primer/probe sequences:
- RBP2-H1::
-
5′-AGTGCAGTGGCGCGATCT-(F),
5′-GGCAGAAGAATTGCTGGAATCTAG-(R),
5′-TGGCTCACTGCAACCT (probe);
- RBBP2H1a::
-
5′-CCCTTGAGTAACTCGCCTTCTGTA-(F),
5′-GGCGTACCTGCCTGAGTAGATG-(R),
5′-CCAGCAATGCCAACGG (probe).
Probes were labeled with a 6-FAM dye as a reporter and a nonfluorescent quencher linked to a groove binder. Primers and probes for 18s rRNA served as endogenous controls using the corresponding pre-designed assay (Assay on demand™, Applied Biosystems, Darmstadt, Germany). For each sample, triplicate reactions were performed as described previously.16, 17, 18 Relative expression ratios were calculated using a standard curve generated with Human Reference RNA (Stratagene, La Jolla, CA, USA) that contains pooled RNAs from 11 different tumor cell lines.13
Immunohistochemistry and Antibodies Used for RBP2-H1 Detection
Tissue microarrays (TMAs) and conventional paraffin-embedded tissue sections were deparaffinized and rehydrated according to standard protocols. After inhibition of the endogenous peroxidase activity with hydrogen peroxide, the sections were incubated at 100°C for 10 min with citrate buffer (pH 6.0). After washing with PBS, samples were blocked with SuperBlockTM (Zytomed, Berlin, Germany) and subsequently incubated with a 1:100 dilution of the primary antibody for 30 min at room temperature. For detection of RBP2-H1, a polyclonal protein A purified antibody against a protein fragment spanning 243 aa of RBP2-H1 was produced in cooperation with Fusion Antibodies, Belfast, Northern Ireland (anti-RBP2-H1243). The 243-aa-protein fragment used for immunization comprises the RBP2-H1-specific domain between aa 237 and aa 273. None of the other known splicing-variants (PLU-1, RBBP2H1a) contains this domain. Detection specifity of anti-RBP2-H1243 was proven by Western blotting. Signals from anti-RBP2-H1243-detected bands were confirmed by a second, also RBP2-H1-specific antibody (customized anti-RBP2-H118; Biocarta Europe, Hamburg, Germany, see below). The ZytoChemPlus HRP Anti Broad Spectrum Staining SystemTM (Zytomed, Berlin, Germany) was used for detection according to the manufacturer's protocol. To avoid experimental bias within the set of tumor samples, immunohistochemistry of all examined tissue samples was performed under identical experimental conditions.
Evaluation of RBP2-H1 Expression Using TMAs
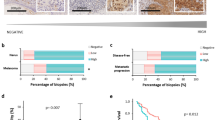
According to the method established by Kononen et al19 three TMAs were designed in cooperation with the Institute of Pathology, University of Regensburg, Germany. TMA ‘No. 1’ represents formalin-fixed paraffin-embedded punches from 52 benign, mainly congenital, melanocytic nevi. On TMA ‘No. 2’, 60 samples from malignant melanomas with different tumor stages are spotted (Table 1). TMA ‘No. 3’ harbors 60 spots from cutaneous and lymph node melanoma metastases. All tissue samples and clinical information are archived at the Department of Dermatology and the Institute of Pathology, University of Regensburg. Immunostaining was assessed by two independent investigators (TV, AR). Inter- and intra-examiner reproducibility was 88 and 92%, respectively. As we have previously described, immunostaining intensity was scored using uniform and clear cutoff criteria to maintain the reproducibility of the method.3 Briefly, the result of immunostaining was recorded as negative or positive considering the expression in tumor cells. Samples were classified as positive if more than 10% of the tumor cells achieved higher signals than the internal positive controls (epidermis, sebaceous glands and vessels).
Evaluation of RBP2-H1 Expression in ‘Whole Tumor’ Sections
Since staining results of the punched samples arrayed on TMAs may be biased by intratumoral heterogeneity of RBP2-H1 expression, also complete tumor sections were analyzed in a second set of experiments (five common nevi, four advanced nodular melanomas, five melanoma metastases, five cutaneous neurofibromas and six cutaneous schwannomas). Since it became apparent that the number of stained cells and the staining intensity can vary within one tumor, a more complex evaluation algorithm was applied for these complete sections according to an algorithm recently described.3 Briefly, to normalize inter-regional heterogeneity between the single tumor samples, three fields of vision were evaluated per section (× 400 magnification). The number of positive stained nuclei was estimated as percentage of all nuclei (p). In addition, the staining intensities (i) of positive cells were scaled. (i) was assessed from grade 0 (no staining) to grade 4 (strong staining, similar to positive control signal). An expression score (ES) was calculated by subsuming mean staining percentages (p) and intensities (i) of all samples (n) of one entity according to the following formula:

where  is the mean percentage of stained nuclei of three representative fields of tumor X,
is the mean percentage of stained nuclei of three representative fields of tumor X,  the mean staining intensity of three representative fields of tumor X, ES the expression score.
the mean staining intensity of three representative fields of tumor X, ES the expression score.
Co-Immunoprecipitation of pRb/RBP2-H1
A375-SM- and IGR-1 melanoma cells as well as a control cell line that expresses high levels of RBP2-H1 (MCF-7) were lysed using Clontech's Extraction/Labeling buffer (Palo Alto, CA, USA). According to standard protocols, immunoprecipitation was performed with protein A sepharose-coupled anti-pRb (4H1) antibodies. By Western blot analyses, co-precipitated RBP2-H1 was detected independently by two different antibodies. (i) A peptide antibody raised against aa 258–275 (customized anti-RBP2-H118; Biocarta Europe, Hamburg, Germany) and (ii) an antibody raised against a 243-aa protein fragment of RBP2-H1 (customized anti-RBP2-H1243; Fusion Antibodies, Belfast, Northern Ireland; details are given above).
Statistical Analysis
The SPSS 10.0. (SPSS Inc., Erkrath, Germany) software package was used to perform statistical analysis. For assessment of relative mRNA expression levels the nonparametric Mann–Whitney U-test, for assessment of intratumoral differences in protein expression the Wilcoxon test for coupled samples was applied. For all statistical tests a probability of error <0.05 was required.
Results
Current Sequence-Based Phylogenetic Analysis of RBPs
Aligning current gene bank entries of known RBPs, a phylogenetic analysis of RBP2-H1-related genes was performed (Figure 1). Some members of the superfamily of RBPs (in total more than 100 members, also including well-studied factors such as E2F, MDM2 or c-Myc) have an intersection with the ARID (AT rich interactive domain) family of DNA-binding proteins characterized by homeodomains such as dri (dead ringer protein, firstly described in Drosophila melanogaster). RBBP2H1a, PLU-1 and RBP2-H1 represent three alternative splicing variants. RBP2-H1 is distinguished by one extra exon of 108 bp. The RBBP2H1a transcript has a much longer 5′-UTR and an ATG upstream of that used for RBP2-H1 and PLU-1. On cDNA level, RBP2-H1 shows a 98.5% identity to PLU-1 and a 98.0% identity to RBBP2H1a. The comparison of RBP2-H1 with RBP-2 reveals a cDNA homology of 37.8%. On protein level, 54.0% of the amino acids of RBP2-H1 and RBP-2 are identical. Sequence analysis of the three splicing variants revealed further highly conserved functional motifs. The dri motif, five nuclear localization motifs, a try/tyr/phe/cys motif and a cys/his motif strongly suggest additional functions of the corresponding proteins as transcription factors.7
Deficiency of RBP2-H1 mRNA in Malignant Melanocytic Tumors
Since previous work has gained preliminary evidence that RBP2-H1 mRNA can be downregulated in UV-irradiated melanocytes and possibly in malignant melanomas,5 we investigated mRNA expression of RBP2-H1 by real-time TaqManTM RT-PCR (Figure 2). Compared to melanocytic nevi (mean 0.29, range 0.13–0.48, s.d.±0.13), RBP2-H1 mRNA was not significantly decreased in melanomas (mean 0.19, range 0.04–0.40, s.d.±0.15) but significantly decreased in melanoma metastases (mean 0.15, range 0.07–0.23, s.d.±0.07; P<0.05). The splicing variant RBBP2H1a also showed the most prominent expression in benign melanocytic tumors. There was a similar, but not significant trend to a deficient expression in melanomas and metastases. Interestingly, similar to melanomas and melanoma metastases, glioblastomas, used as external controls representing another neuroectodermal cancer, also showed a relative loss of RPB2-H1 compared to fetal brain. In contrast, epithelial cancers, in particular breast cancer, showed a comparably high RBP2-H1 expression and only marginal RBBP2H1a expression.
Relative quantitation of RBP2-H1 and RBBP2H1a expression in neuroectodermal tumors in contrast to non-neuroectodermal controls. Analysis of microdissected tissue samples from six benign melanocytic nevi, five advanced nodular melanomas and seven melanoma metastases revealed a progressive deficiency in melanocytic tumors. Samples from five glioblastomas, used as external control representing another neuroectodermal cancer, also showed a relative loss of RPB2-H1 compared to fetal brain. In accordance with previous studies higher expression levels are sustained in epithelial cancers. Error bars indicate standard deviations (ns, not significant).
Immunohistochemical Staining Confirms the Deficiency of RBP2-H1 in Malignant Melanocytic Tumors
The progressive deficiency of RBP2-H1 mRNA from common nevi to melanoma and melanoma metastasis could be significantly confirmed on protein level. TMAs were examined to evaluate gross constitutive differences in RBP2-H1-expression between benign melanocytic lesions and malignant melanocytic tumors (Figure 3). In accordance to our RT-PCR data, TMA-based immunohistochemical analyses confirmed a progressive deficiency of RBP2-H1 expression from nevi (TMA No. 1, n=52) to melanoma (TMA No. 2, n=60) and melanoma metastases (TMA No. 3, n=60). Almost 70% of the spotted nevus samples exceeded the threshold of 10% positive stained cells (see Materials and methods) and, therefore, were classified as ‘RBP2-H1-expressors’. Strikingly, in melanomas, RBP2-H1 staining was only detected in primary radial growth phase melanomas with a cutoff tumor thickness of ≤1.6 mm (Table 1). In this group, 31.6% were positive, which adds up to only 10% of all melanomas spotted on TMA No. 2. In thicker more advanced melanomas (>1.6 mm) no staining signals could be found. However, in melanoma metastases, 30% of the samples did express RBP2-H1.
Owing to limited punch size and intratumoral heterogeneity in TMAs, a methodical bias is possible. Therefore, an additional analysis of complete tissue sections was performed, too. With regard to the whole tumor architecture, no region-specific distribution (eg tumor front or tumor core) of RBP2-H1 could be detected. However, a certain heterogeneity of staining results (number, intensity) across the full sections could be seen. Therefore, an appropriate scoring system was applied (see Materials and methods). In accordance with our mRNA measurements and the TMA analyses, the highest RBP2-H1 expression scores were found in nevi with a significant decrease in both melanomas (P<0.03) and metastases (P<0.02), respectively. Other benign neuroectodermal tumors that served as independent controls such as schwannomas and neurofibromas showed a comparably high ‘melanocytic nevus-like’ RBP2-H1-expression (Table 2; Figure 4a–c).
RBP2-H1 Expression in Cell Lines
Real-time TaqManTM RT-PCR revealed heterogeneous RBP2-H1 expression patterns among the different melanoma cell lines Mel-HO, IGR-1, A375-SM and COLO-849 (Figure 5a). As control, the breast cancer cell line MCF-7 was used, because in the tissue analysis shown above, breast cancer reached a relatively high RBP2-H1 expression level similar to that of common melanocytic nevi. In Western blots using the specifically generated anti-RBP2-H118 antibody, a weak correlation of RBP2-H1 protein expression with RBP2-H1 transcript levels detected by RT-PCR was seen (Figure 5b). As an exception, COLO-894 expressed high RBP2-H1 levels in both RT-PCR and Western blotting. Interestingly, A375-SM cells characterized as strongly metastatic14 revealed an intermediate RNA expression and a low protein expression.
Relative quantitation of RBP2-H1 expression in different melanoma cell lines Mel-HO, IGR-1, A375-SM and COLO-849 and in MCF-7, a breast cancer-derived cell line as control. Error bars indicate standard deviations (a). Western blot analysis of RBP2-H1 expression in different melanoma cell lines and in MCF-7, a breast cancer-derived cell line as control. For detection a specifically generated anti-RBP2-H118 antibody was used. Equal protein amounts (40 μg) were loaded on each lane (b).
RBP2-H1 Co-Precipitates with pRb
Since RBP-2 was previously suggested to be involved in pRb-phosphorylation control by direct protein–protein interaction,4 we addressed the question whether RBP2-H1, the closest homolog of RBP-2, could also bind to pRb. While RBP2-H1 lacks the typical Leu-X-Cys-X-Glu-motif of some pRb-interacting proteins, it bears a homolog-domain of the non-T/E1A-pRb-binding domain of RBP-2.5 The latter could theoretically confer a direct interaction with pRb and make RBP2-H1 a true pRb-binding protein.
To evaluate this possibility, co-immunoprecipitation studies were performed with lysates from IGR-1 and A375-SM melanoma cells and, as control, MCF-7 breast cancer cells. In both, IGR-1 and MCF-7, after co-immunoprecipitation with anti-pRb 4H1 antibodies, a specific band at about 160 kDa was detected by the specifically generated RBP2-H1 antibody (anti-RBP2-H118, Figure 6). The same band could also be detected by the second RBP2-H1-specific antibody (protein fragment antibody anti-RBP2-H1243), which was raised against an epitope of RBP2-H1 different from that of anti-RBP2-H118 (not shown). A375-SM failed to show a significant co-immunoprecipitation probably due to their relative deficit in RBP2-H1 protein expression (Figure 5b). Our results also demonstrate that only a subfraction of total RBP2-H1, seen in the cell lysate and supernatant, is bound to pRb suggesting a possible balance between bound and unbound RBP2-H1.
Co-immunoprecipitation studies reveal an association of RBP2-H1 with pRb in IGR-1 melanoma cells and a control cell line (MCF-7) that expresses high levels of RBP2-H1. A375-SM melanoma cells failed to show detectable co-immunoprecipitation possibly due to low RBP2-H1 expression. For immunoprecipitation the anti-pRb antibody Rb (4H1) and for detection of RBP2-H1 two independent RBP2-H1-specific antibodies were successively used (anti-RBP2-H118 depicted, anti-RBP2-H1243 not shown). Equal protein amounts (40 μg) were loaded on each lane.
Discussion
Our study demonstrates that RBP2-H1, a protein with high sequence homology to members of the family of RBPs, is progressively decreased in malignant melanocytic tumors. The observed deficiency seems to be quite specific for neuroectodermal derived cancers (melanomas and glioblastomas) since RBP2-H1 seems to be abundantly expressed in epithelial cancers. In accordance with our RT-PCR measurements, Barrett et al7 reported an upregulation of PLU-1 in breast cancer. PLU-1 and RBBP2H1a represent splicing variants of RBP2-H1.
On mRNA level, the deficiency of RBP2-H1 was only significant in melanoma metastases, but not in primary melanomas. This may be explained by the limited number of samples in the RNA analysis, since the TMA–immunohistochemistry data prove a deficit of RBP2-H1 expression already in primary melanomas. Interestingly, the immunohistochemically stained ‘whole tumor’ samples revealed varying protein expression within the single sections, suggesting the possible existence of cell clones with differential RBP2-H1 expression within a single tumor. Accordingly, the varying RBP2-H1 expression levels seen in melanoma cell lines may be explained by tumor heterogeneity.
Considering our TMA results, the question may arise, why in the ‘melanoma TMA No. 2’ RBP2-H1 expression dropped down to 10% positive stained samples, whereas in the following ‘metastasis TMA No. 3’ again an expression was found in 30% of the spotted samples. A possible reason could be that on the ‘melanoma TMA’ just 19 samples with a tumor thickness ≤1.6 mm have been spotted. This means just about one-third of all spotted samples represents potential staining candidates, at all. If we would have created a TMA with 60 samples of exclusively thin melanomas, probably, a higher percentage of stained spots would have been detected. Moreover, in contrast to the spotted samples from thick melanomas on TMA No. 2, in the evaluation of ‘whole tumor sections’, we also detected RBP2-H1 expression in some advanced nodular melanomas. Hence, the relatively low counts in primary melanoma are probably due to the common ‘keyhole’ problem of TMAs. In any case, the loss of RBP2-H1 expression seems to be correlated with the tumor thickness and the progression of malignant melanomas. Although no statement can be made concerning a causal role or contribution to progressive decrease, RBP2-H1 is a candidate progression marker for melanomas. A further clinicopathological evaluation of the RBP2-H1 status in melanocytic tumors could provide new insights into the possible impact of RBP2-H1 deficiency also on melanoma prognosis. Moreover, RBP2-H1 would deserve a more detailed molecular investigation if a direct binding to pRb, the central controller of the cell cycle, seemed likely.
Therefore, we used the established antibodies also for co-immunoprecipitation studies. A pRb-binding activity of RBP2-H1 has been recently hypothesized only on the basis of sequence alignments with other RBPs, namely RBP-2.5 However, a protein–protein interaction has not yet been demonstrated. Using lysates from IGR-1 and MCF-7 cells, we can now demonstrate that a subfraction of total RBP2-H1 is associated with pRb making RBP2-H1 a true pRb-binding protein with possible functional impact on pRb. On the other hand, since supernatants harbored considerable amounts of RBP2-H1, possibly a balance between bound and unbound RBP2-H1 exists.
The detected pRb/RBP2-H1-association in co-immunoprecipitation can either be due to a colocalization of RBP2-H1 with pRb within a multimolecular protein complex or to a direct RBP2-H1/pRb-binding. Members of the pRb family (pRb/p105, p107 and p130) were initially reported to interact with various proteins via the characteristic A/B pocket domain. Binding partners, such as the viral oncoproteins E1A, large T and E7, contain a Leu-X-Cys-X-Glu docking site, which is responsible for a stable interaction with several residues within the pocket. Furthermore, other Leu-X-Cys-X-Glu-independent ways of an interaction with pRb have been described.20, 21, 22 For example, non-T/E1A-pRb-binding domains are located in the C-terminal region of both RBP-2 and a corresponding homolog region in RBP2-H1 (RBP-2: aa 1453–1563, RBP2-H1 aa 1391–1490).5, 11 Since RBP-2 binds and probably stabilizes active, hypophosphorylated pRb, also for other RBPs, inclusively RBP2-H1, an impact on pRb phosphorylation status and pRb/E2F-mediated cell cycle control is conceivable.11, 12
The four melanoma cell lines Mel-HO, IGR-1, A375-SM and COLO-849 were chosen for expression analysis in our study because they are characterized by different doubling times (supplied by the providers). Considering proliferation rates, however, no correlation to the respective RBP2-H1 expression could be seen for Mel-HO, IGR-1 and COLO-849. However A375-SM cells, which are characterized by a strong metastatic potential,14 exhibited very low protein expression of RBP2-H1. Therefore, A375-SM could serve as a model cell line for studying functional consequences of RBP2-H1 re-expression in the future.
We conclude that RBP2-H1 is progressively downregulated in malignant melanocytic tumors. As a direct interaction with pRb seems possible, a causal contribution of RBP2-H1 deficiency to melanoma progression can be envisioned.
Accession codes
References
Bartkova J, Lukas J, Guldberg P, et al. The p16-cyclin D/Cdk4-pRb pathway as a functional unit frequently altered in melanoma pathogenesis. Cancer Res 1996;56:5475–5483.
Halaban R, Miglarese MR, Smicun Y, et al. Melanomas, from the cell cycle point of view (Review). Int J Mol Med 1998;1:419–425.
Roesch A, Becker B, Meyer S, et al. Overexpression and hyperphosphorylation of retinoblastoma protein in the progression of malignant melanoma. Mod Pathol 2004, Epub ahead of print.
Defeo-Jones D, Huang PS, Jones RE, et al. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature 1991;352:251–254.
Vogt T, Kroiss M, McClelland M, et al. Deficiency of a novel retinoblastoma binding protein 2-homolog is a consistent feature of sporadic human melanoma skin cancer. Lab Invest 1999;79:1615–1627.
Kashuba V, Protopopov A, Podowski R, et al. Isolation and chromosomal localization of a new human retinoblastoma binding protein 2 homologue 1a (RBBP2H1A). Eur J Hum Genet 2000;8:407–413.
Barrett A, Madsen B, Copier J, et al. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen? Int J Cancer 2002;101:581–588.
Lu PJ, Sundquist K, Baeckstrom D, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem 1999;274:15633–15645.
Tan K, Shaw AL, Madsen B, et al. Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J Biol Chem 2003;278:20507–20513.
Meehan WJ, Samant RS, Hopper JE, et al. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem 2004;279:1562–1569.
Fattaey AR, Helin K, Dembski MS, et al. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 1993;8:3149–3156.
Kim YW, Otterson GA, Kratzke RA, et al. Differential specificity for binding of retinoblastoma binding protein 2 to RB, p107, and TATA-binding protein. Mol Cell Biol 1994;14:7256–7264.
Hafner C, Bataille F, Meyer S, et al. Loss of EphB6 expression in metastatic melanoma. Int J Oncol 2003;23:1553–1559.
Gehlsen KR, Davis GE, Sriramarao P . Integrin expression in human melanoma cells with differing invasive and metastatic properties. Clin Exp Metastasis 1992;10:111–120.
Hafner C, Schmitz G, Meyer S, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem 2004;50:490–499.
Rumpler G, Becker B, Hafner C, et al. Identification of differentially expressed genes in models of melanoma progression by cDNA array analysis: SPARC, MIF and a novel cathepsin protease characterize aggressive phenotypes. Exp Dermatol 2003;12:761–771.
Tang XX, Zhao H, Robinson ME, et al. Prognostic significance of EPHB6, EFNB2, and EFNB3 expressions in neuroblastoma. Med Pediatr Oncol 2000;35:656–658.
Tang XX, Evans AE, Zhao H, et al. Association among EPHB2, TrkA, and MYCN expression in low-stage neuroblastomas. Med Pediatr Oncol 2001;36:80–82.
Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–847.
Kaelin Jr WG . Recent insights into the functions of the retinoblastoma susceptibility gene product. Cancer Invest 1997;15:243–254.
Taya Y . RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci 1997;22:14–17.
Stiegler P, Giordano A . The family of retinoblastoma proteins. Crit Rev Eukaryot Gene Expr 2001;11:59–76.
Acknowledgements
The technical assistance of Mrs Lydia Kuenzel and Nadine Wandtke is gratefully acknowledged. This work was funded by the Deutsche Forschungsgemeinschaft, DFG-Grant: KR 2135/1-1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roesch, A., Becker, B., Meyer, S. et al. Retinoblastoma-binding protein 2-homolog 1: a retinoblastoma-binding protein downregulated in malignant melanomas. Mod Pathol 18, 1249–1257 (2005). https://doi.org/10.1038/modpathol.3800413
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800413
Keywords
This article is cited by
-
A truncated and catalytically inactive isoform of KDM5B histone demethylase accumulates in breast cancer cells and regulates H3K4 tri-methylation and gene expression
Cancer Gene Therapy (2023)
-
Persister state-directed transitioning and vulnerability in melanoma
Nature Communications (2022)
-
The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism
Experimental & Molecular Medicine (2019)
-
Prognostic significance of RBP2-H1 variant of JARID1B in melanoma
BMC Cancer (2017)
-
Targeting the epigenome for treatment of cancer
Oncogene (2012)