Abstract
The p16INK4A/CDKN2A (p16) gene on chromosome 9p21 is inactivated in >90% of invasive pancreatic cancers. In 40% of pancreatic cancers the p16 gene is inactivated by homozygous deletion, in 40% by an intragenic mutation coupled with loss of the second allele, and in 10–15% by hypermethylation of the p16 gene promoter. Immunohistochemical labeling for the p16 gene product parallels gene status, but does not provide information of the mechanism of p16 gene inactivation. The methylthioadenosine phosphorylase gene (MTAP) gene also resides on chromosome 9p21, approximately 100 kb telomeric to the p16 gene. The MTAP gene is frequently contained within p16 homozygous deletions, producing concordant loss of both p16 and MTAP gene expression. Concordant loss of both p16 and MTAP protein expression can therefore be used as a surrogate marker for p16 homozygous deletion. Here we immunolabeled a series of pancreatic intraepithelial neoplasia (PanIN) lesions of various histologic grades for the p16 and MTAP gene products using a high-throughput PanIN tissue microarray (TMA) format. We demonstrate concordant loss of p16 and MTAP protein expression in 6/73 (8%) PanINs, including five high-grade lesions and one low-grade lesion. Immunolabeling for both p16 and MTAP protein expression provides a tool to evaluate tissues with intact morphology for p16 gene homozygous deletions. The concordant loss of expression of both genes in PanIN lesions demonstrates that homozygous deletions of the p16 tumor suppressor gene can occur in noninvasive precursor lesions.
Similar content being viewed by others
Main
Pancreatic intraepithelial neoplasia (PanIN) is a powerful system to study noninvasive precursors of an infiltrating cancer. First, the disease is important. The infiltrating cancer associated with PanINs, infiltrating adenocarcinoma of the pancreas, is the fourth leading cause of cancer death.1 This year approximately 31,000 Americans will be diagnosed with PanINs infiltrating adenocarcinoma of the pancreas and 31 000 will die from it. Second, PanINs are histologically well-defined. An international consensus has been developed for the classification and grading of PanINs, allowing investigators at one institution to compare their results directly with findings from another institution.2 Third, the genetics of PanINs are well described, and a progression model for the occurrence of genetic alterations in PanINs has been developed.3 Telomere shortening and activating point mutations in the KRAS2 oncogene occur early in PanIN-1 lesions, the p16INK4A/CDKN2A (henceforth referred to as p16) gene is inactivated in intermediate and late lesions (PanINs 2 and 3), and the TP53, MADH4, and BRCA2 genes are inactivated late, in PanIN-3 lesions.4, 5, 6, 7, 8, 9, 10
One of the most important of the genetic alterations in infiltrating adenocarcinoma of the pancreas and in PanIN, as judged by its high prevalence, is inactivation of the p16 gene. The p16 gene is inactivated in 40% of pancreatic cancers by homozygous deletion, in 40% by an intragenic mutation coupled with loss of the second allele, and in 15% by hypermethylation of the p16 gene promoter.11, 12, 13 Immunohistochemical labeling has demonstrated that 30% of PanIN-1 lesions, 55% of PanIN-2 lesions, and 70% of PanIN-3 lesions show loss of expression of the p16 gene product.14 The mechanism of this loss of expression in PanINs is, however, not well-defined. In specific, because almost pure samples are required to detect homozygous deletions by analysis of DNA (even a small amount of normal tissue may obscure the deletion), it is extremely difficult to detect homozygous deletions in tissue sections. The ‘tissue architecture’ of homozygous deletions of the p16 gene in infiltrating adenocarcinoma of the pancreas and its precursors remains poorly defined.
Immunolabeling for both the MTAP and p16 gene products provides a unique opportunity to define the architecture of p16 homozygous deletions in tissue sections. The MTAP gene is located approximately 100 kb telomeric to the p16 gene on chromosome 9p21.15 The MTAP gene is contained in the p16 homozygous deletion in 90% of malignant mesotheliomas, in 75–100% of T-cell acute lymphoblastic leukemias, and in 50–100% of pancreatic cancers.16, 17, 18, 19 Importantly, immunolabeling for the MTAP protein closely mirrors gene status. For example, our group has recently demonstrated that loss of MTAP expression in pancreatic cancers is present in only a subset of cases with known p16 gene homozygous deletions, specifically that subset where the 9p21 deletion is large enough to encompass the MTAP gene.20 Secondly, using a high-throughput microarray approach, our group studied a series of >440 pancreatic and periampullary cancers, and found that loss of MTAP expression in these cancers always occurs in conjunction with loss of p16 protein expression,20 confirming that immunolabeling for the MTAP protein can be used as a surrogate marker for homozygous deletions of the p16 gene.
In this study, we extend this unique opportunity for determining mechanism of gene inactivation to preneoplastic lesions of pancreatic cancer, and demonstrate concordant loss of both p16 and MTAP gene expression in a series of PanIN lesions, thereby establishing the existence of homozygous deletions in a noninvasive precursor to an invasive cancer.
Materials and methods
Tissues
The study was approved by the Johns Hopkins Institutional Review Board. Paraffin-embedded tissue blocks of pancreatectomy specimens were retrieved from the Surgical Pathology archives of the Johns Hopkins Hospital; all blocks were retrieved from cases that were resected for ductal adenocarcinoma of the pancreas. PanIN lesions were identified in the adjacent pancreata by two pathologists extensively familiar with PanIN morphology (RHH and AM), and tissue microarrays (TMAs) were created from these lesions as previously described.9 Two TMA slides encompassing 90 individual PanIN lesions and a variety of non-neoplastic, nonpancreatic organs were constructed. The PanINs were graded using an internationally established nomenclature and grading scheme.2 In order to simplify analyses, PanINs-1A and 1B were combined into ‘low-grade’ and PanINs-2 and 3 into ‘high-grade’, as we have previously described.7
MTAP Labeling
We utilized a newly developed immunohistochemical assay for the detection of absence of MTAP expression using a novel monoclonal anti-MTAP antibody developed by GeneTex Inc., San Antonio, TX, USA and Salmedix, Inc., San Diego, CA, USA. Unstained 4 μm sections were cut from each TMA block and deparaffinized by routine techniques. Antigen retrieval was performed by incubating the tissue-sections in Borg Decloaker at 120°C (Biocare Medical, CA, USA) for 3 min, followed by trypsin incubation for 5 min at room temperature. The sections were incubated with 20 μg/ml of the monoclonal antihuman MTAP antibody (Clone 6.9, Salmedix, Inc., San Diego, CA, USA). Incubation of labeled polymer (Dako, Envision Plus Detection Kit, Carpinteria, CA, USA) was carried out for 30 min at room temperature. The peroxidase reaction was visualized by incubating with DAB (3,3′-diaminobenzidine tetrahydrochloride) for 5 min.
p16 Labeling
Serial 4 μm paraffin sections were obtained from the two TMA blocks and deparaffinized by routine techniques before they underwent a 38 min pretreatment using Ventana Cell Conditioner 1 (Ventana Medical Systems, Inc., Tucson, AZ, USA) after the slides had been warmed up for 8 min at 95°C and for 4 min at 100°C. The slides where quenched with 3% H2O2 for 4 min before incubating with a mouse monoclonal antihuman p16 antibody in its prediluted ready to use form (Clone 16P04, Catalog No.CMA811, Cell Marque Corp., Hot Springs, AR, USA) for 28 min using the automated Ventana Benchmark XT (Ventana Medical Systems, Inc., Tucson, AZ, USA). Labeling was detected with the iVIEW™ DAB Detection Kit (Ventana Medical Systems, Inc., Tucson, AZ, USA) as per the manufacturer's protocol. All sections were counterstained with Giles’ hematoxylin and blued in Ventana Bluing Reagent™.
Grading of Labeling
Immunolabeling was evaluated by three investigators (SH, RHH, AM) at a multiheaded microscope with consensus reached in all cases. Immunolabeling for the p16 gene product was evaluated as has been described. Complete loss of nuclear labeling was considered negative, while any nuclear labeling was considered positive. The immunolabeling for the MTAP gene product was interpreted in a similar fashion. Complete loss of cytoplasmic labeling was considered negative, while any labeling was considered positive.
Results
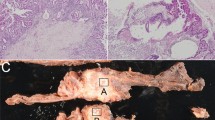
Of the 90 PanIN lesions arrayed on the two TMA slides, 73 (81%) were appropriate for evaluation for both MTAP and p16 immunohistochemical labeling. The attrition occurred primarily due to ‘missing’ the intraductal lesion during array construction, and occasionally due to tissue artifacts. The 73 PanIN lesions included 41 (56%) low-grade lesions and 32 (44%) high-grade lesions. Robust MTAP expression was seen in 67/73 (90%) PanINs, and when present, MTAP labeling almost always involved the entire circumference of the intraductal lesion (Figure 1a). By contrast, complete loss of MTAP labeling was seen in 6/73 (8%) of PanIN lesions, and included five high-grade and one low-grade PanIN. Loss of MTAP expression involved all neoplastic cells within the PanIN lesion in the six cases (Figure 1b), and adjacent non-neoplastic ductal epithelium and/or acinar tissue were generally available within the TMA core to ensure adequacy of staining. Six of six (100%) of PanINs with loss of MTAP expression also demonstrated loss of p16 labeling, that is, loss of MTAP labeling with retained p16 expression was not seen in our series of PanIN lesions. Of note, one PanIN-3 lesion demonstrated loss of MTAP labeling in a portion of the epithelium, with an abrupt transition to a segment with intact expression (Figure 1c). On the serial section, retained p16 labeling was present in the identical region of the epithelium, suggesting a homozygous loss of both genes within a portion of the precursor lesion (Figure 1d). Interestingly, the morphologic appearance of the segment of the PanIN-3 with loss of p16 and MTAP was similar to the morphologic appearance of focus of the PanIN-3 with intact p16 and MTAP expression. This single PanIN-3 lesion with ‘focal’ p16 and MTAP expression was excluded from the final analysis.
(a) Intact MTAP labeling in a high-grade PanIN lesion. (b) Loss of MTAP labeling in a high-grade PanIN lesion. Note retained expression in subjacent normal pancreatic ductal epithelium. (c) Partial loss of MTAP expression in a PanIN lesion with retained expression within a portion of the duct (arrow), (d) Serial section of tissue microarray demonstrates loss of p16 labeling with retained nuclear labeling within the identical focus (arrow) in the PanIN lesion.
Discussion
In this study, we take advantage of the frequent codeletion of two genes to develop a simple immunolabeling technique to identify sites of homozygous deletion in formalin-fixed paraffin-embedded tissue sections. Immunolabeling for the MTAP protein can be used as a surrogate marker for homozygous deletions of the p16 gene because the MTAP gene is frequently included in the p16 homozygous deletion, and because MTAP protein expression closely correlates with MTAP gene status.16, 19, 20, 21 In addition, when immunolabeling for the p16 gene product is combined with immunolabeling for MTAP, the loss of labeling for both MTAP and the p16 gene product in the exact same lesions or portions of lesions supports a common mechanism of inactivation for these two genes.
The concordant loss of both p16 and MTAP gene expression, as found here in PanIN lesions, essentially demonstrates that a homozygous deletion in a tumor suppressor gene had occurred in a noninvasive precursor lesion to invasive pancreatic cancer. This conclusion is analogous to previous molecular analyses of PanIN lesions demonstrating biallelic inactivation by a combination of hemizygous deletion and intragenic mutation,5, 6 and adds to the growing body of literature that illustrates a progression of genetic alterations from morphologically normal ductal epithelial cells, to PanIN lesions, to infiltrating ductal adenocarcinoma.2, 4, 8
The immunolabeling approach described here has several advantages over formal molecular genetic analyses. The latter analyses do not preserve tissue histology and therefore may not detect topographically focal deletions of the p16 gene, as they rely on the microdissection of morphologically identifiable lesions.22, 23 These microdissections are likely to comingle cells having different p16 gene status in instances where the cells have an identical morphologic appearance. Microdissection can therefore obscure deletions that do not involve an entire morphologically defined lesion. In addition, molecular analyses of DNA samples for p16 homozygous deletions in tissue sections are extremely time consuming and cumbersome. By contrast, the approach here can be performed in any routine immunohistochemistry lab.
Additional potential applications of the technique presented here include: (1) the analyses of precursor lesions and invasive cancers of other organs, and (2) the analyses of metachronous carcinomas to determine whether they represent a primary and a metastasis, or two independent primary tumors. For example, the finding of a homozygous deletion of the p16 gene in a patient's first carcinoma and intact p16 in their second carcinoma would provide strong evidence that the second neoplasm was an independent second primary.
Although the immunolabeling approach presented here can define some patterns of p16 gene homozygous deletions in tissue sections, it has several limitations that should be acknowledged. First, depending on the size of the homozygous deletion, some homozygous deletions of the p16 gene will not encompass the MTAP gene. For example, the MTAP gene is not contained in the p16 homozygous deletion in ∼10% of malignant mesotheliomas.19 Therefore, a fraction of 10% of p16 homozygous deletions would be missed by immunolabeling studies of the MTAP gene product. Conversely, although there is a very high correlation between MTAP protein expression and MTAP gene status, homozygous deletion of both genes may not be the only mechanism by which the MTAP gene is inactivated. Although these alternate mechanisms of MTAP gene inactivation (eg, promoter methylation) are theoretically possible, our data on >440 pancreatic and periampullary cancers suggest that this appears to be essentially absent in the pancreas.20
In summary, combined immunolabeling for the MTAP and p16 gene products can be used as a surrogate marker for homozygous deletions of the p16 gene in tissue sections. Here we use this approach to demonstrate the homozygous deletion of the p16 gene in a series of noninvasive precursor lesions to an invasive cancer.
References
Yeo TP, Hruban RH, Leach SD, et al. Pancreatic cancer. Curr Probl Cancer 2002;26:176–275.
Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol 2004;28:977–987.
Hruban RH, Wilentz RE, Kern SE . Genetic progression in the pancreatic ducts. Am J Pathol 2000;156:1821–1825.
Biankin AV, Kench JG, Dijkman FP, et al. Molecular pathogenesis of precursor lesions of pancreatic ductal adenocarcinoma. Pathology 2003;35:14–24.
Goggins M, Hruban RH, Kern SE . BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am J Pathol 2000;156:1767–1771.
Luttges J, Galehdari H, Brocker V, et al. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol 2001;158:1677–1683.
Maitra A, Adsay NV, Argani P, et al. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol 2003;16:902–912.
Takaori K, Hruban RH, Maitra A, et al. Pancreatic intraepithelial neoplasia. Pancreas 2004;28:257–262.
van Heek NT, Meeker AK, Kern SE, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol 2002;161:1541–1547.
Wilentz RE, Iacobuzio-Donahue CA, Argani P, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res 2000;60:2002–2006.
Caldas C, Hahn SA, da Costa LT, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet 1994;8:27–32.
Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res 1997;57:3126–3130.
Geradts J, Hruban RH, Schutte M, et al. Immunohistochemical p16INK4a analysis of archival tumors with deletion, hypermethylation, or mutation of the CDKN2/MTS1 gene. A comparison of four commercial antibodies. Appl Immunohistochem Mol Morphol 2000;8:71–79.
Wilentz RE, Geradts J, Maynard R, et al. Inactivation of the p16 (INK4A) tumor-suppressor gene in pancreatic duct lesions: loss of intranuclear expression. Cancer Res 1998;58:4740–4744.
Olopade OI, Pomykala HM, Hagos F, et al. Construction of a 2.8-megabase yeast artificial chromosome contig and cloning of the human methylthioadenosine phosphorylase gene from the tumor suppressor region on 9p21. Proc Natl Acad Sci USA 1995;92:6489–6493.
Chen ZH, Zhang H, Savarese TM . Gene deletion chemoselectivity: codeletion of the genes for p16(INK4), methylthioadenosine phosphorylase, and the alpha- and beta-interferons in human pancreatic cell carcinoma lines and its implications for chemotherapy. Cancer Res 1996;56:1083–1090.
Harasawa H, Yamada Y, Kudoh M, et al. Chemotherapy targeting methylthioadenosine phosphorylase (MTAP) deficiency in adult T cell leukemia (ATL). Leukemia 2002;16:1799–1807.
Efferth T, Miyachi H, Drexler HG, et al. Methylthioadenosine phosphorylase as target for chemoselective treatment of T-cell acute lymphoblastic leukemic cells. Blood Cells Mol Dis 2002;28:47–56.
Illei PB, Rusch VW, Zakowski MF, et al. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res 2003;9:2108–2113.
Hustinx SR, Hruban RH, Leoni LM, et al. Homozygous deletion of the MTAP gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapy. Cancer Biol Ther 2005;4:83–86.
Nobori T, Takabayashi K, Tran P, et al. Genomic cloning of methylthioadenosine phosphorylase: a purine metabolic enzyme deficient in multiple different cancers. Proc Natl Acad Sci USA 1996;93:6203–6208.
Maitra A, Wistuba II, Gazdar AF . Microdissection and the study of cancer pathways. Curr Mol Med 2001;1:153–162.
Maitra A, Gazdar AF . Tissue microdissection and processing. Cancer Treat Res 2001;106:63–84.
Acknowledgements
This work was supported by the NIH SPORE (Specialized Programs of Research Excellence) in Gastrointestinal Cancer Grant CA62924, the Michael Rolfe Foundation for Pancreatic Cancer Research, and the Sol Goldman Center for Pancreatic Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hustinx, S., Leoni, L., Yeo, C. et al. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod Pathol 18, 959–963 (2005). https://doi.org/10.1038/modpathol.3800377
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800377
Keywords
This article is cited by
-
Dominant role of CDKN2B/p15INK4B of 9p21.3 tumor suppressor hub in inhibition of cell-cycle and glycolysis
Nature Communications (2021)
-
Regulation of metabolic reprogramming by tumor suppressor genes in pancreatic cancer
Experimental Hematology & Oncology (2020)
-
Systems Oncology: Bridging Pancreatic and Castrate Resistant Prostate Cancer
Pathology & Oncology Research (2019)
-
Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions
Nature Communications (2017)
-
Expression and clinical significance of IMP3 in microdissected premalignant and malignant pancreatic lesions
Clinical and Translational Oncology (2015)