Abstract
Array-based comparative genomic hybridization (CGH) is a technique that allows genome wide screening of gains and losses in DNA copy number. In cases where multiple tumors are encountered, this genetic technique may prove useful in differentiating new primary tumors from recurrences. In this case report, we used array-based CGH to examine the genomic relationships among two leiomyosarcomas and two breast cancers in the same patient, three of which were diagnosed synchronously. Array-based CGH was performed on the four tumor samples using random prime amplified microdissected DNA. Samples were hybridized onto bacterial artificial chromosome arrays composed of approximately 2400 clones. Patterns of alterations within the tumors were compared and genetic alterations among the leiomyosarcomas and breast lesions were found. Overall, three distinct genetic profiles were observed. While the two leiomyosarcomas shared a similar pattern of genetic alterations, the two invasive breast lesions did not. The nearly identical pattern of genetic alterations belonging to the two metachronous leiomyosarcomas confirmed metastatic recurrence while the two different genetic profiles of the invasive ductal carcinomas suggest that the two lesions represented two distinct foci of multifocal disease rather than clonal extension of the primary tumor. We conclude that genetic analysis by array-based CGH can clearly elucidate the relationships between multiple tumors and may potentially serve as an important clinical tool.
Similar content being viewed by others
Main
Comparative genomic hybridization (CGH) is a technique that has greatly facilitated detection of gains and losses in DNA copy number, and has been of particular utility in elucidating the pattern of genomic alterations in tumors. Although this technique remains primarily a research tool, it has potential clinical use in cases with complex presentation. Array CGH may be of clinical value in differentiating new primary tumors from recurrent lesions, particularly in patients with unusual recurrence patterns. Genomic analysis can also help elucidate the relationship between multiple tumor foci. These determinations may in some cases significantly alter the course of further treatment. Array CGH can thus serve as an important adjunct to immunohistochemistry and histopathology. In this study, we examined the genomic relationships between four separate cancers in the same patient, using array CGH. This case illustrates the potential clinical application of genomic technology in defining the relationship between multiple tumors.
Case report
A 43-year old woman 5 years status post resection of a high-grade left retroperitoneal leiomyosarcoma presented with masses in the left breast and right thigh. Mammography showed a highly suspicious spiculated lesion in the lower inner left breast, corresponding to the palpable mass. Fine-needle aspiration of the breast lesion confirmed the diagnosis of adenocarcinoma. Fine-needle aspiration of the thigh lesion was also performed, which demonstrated a spindle cell neoplasm. The patient underwent a modified radical mastectomy of the left breast, and wide excision of the thigh mass. Final pathology demonstrated two separate breast cancers and leiomyosarcoma of the thigh.
Materials and methods
Microdissection and DNA Extraction
Exemption from IRB approval was obtained. Histology of all tumors was reviewed by a pathologist (YYC). DNA extraction and microdissection was performed as previously described.1, 2, 3 Briefly, tumor cells were manually microdissected from a series of seven slides cut from paraffin-embedded archival tumor specimens. The tissue was digested in proteinase K for 3 days and DNA was quantitated by Taqman real-time PCR.
Amplification and Labeling
Approximately 100 ng of tumor DNA and 50 ng of fresh genomic reference DNA (Promega, Madison, WI, USA) was random prime amplified and random prime labeled as previously described using materials from the BioPrime DNA Labeling System (Invitrogen, Carlsbad, CA, USA).3 Following amplification, the tumor DNA was labeled with FluoroLink Cy3-dUTP and reference DNA was labeled with FluoroLink Cy5-dUTP (Amersham Pharmacia, Piscataway, NJ, USA). Unincorporated nucleotides were removed using Sephadex G-50 spin columns (Amersham Pharmacia, Piscataway, NJ, USA).
Array-Based CGH
Array CGH was performed according to protocols described previously.3, 4, 5 To prepare the DNA probe, 80 μl of labeled tumor DNA and 80 μl of labeled reference DNA were combined and mixed with 100 μg Cot-1 DNA (Invitrogen, Carlsbad, CA, USA). The mixture was precipitated with 1/10th 3 M sodium acetate and 2.5 × 100% ethanol. The precipitated DNA was redissolved in a solution of 100 μg/μl yeast t-RNA with 20% SDS. This was then thoroughly mixed with 42 μl of hybridization solution consisting of 71% formamide, 14% dextran sulfate, and 2.9 × SSC. The DNA probe mix was denatured at 70°C for 15 min, and then incubated in 37°C for 30 min. The DNA probe was applied to Human Array 2.0 chromium surface arrays provided by the UCSF Cancer Center Array Core. Each array was composed of 2464 unique sequence bacterial artificial chromosomes printed in triplicate and UV cross-linked at 1200 μJ with a UV stratalinker (La Jolla, CA, USA). After applying the probe, the slide was sealed in a hybridization chamber and incubated upright for 48 h at 37°C on a slowly rocking table.
After hybridization, the slide was washed in 50% formamide/2 × SSC at 48°C for 15 min followed by a wash in 2 × SSC/0.1% SDS solution at 48°C for 20 min. The slide was washed twice in phosphate buffer with 0.1% NP-40, pH=8.0 at room temperature for 10 min. A measure of 100 μl of 3 μg/ml DAPI in 10% PBS in glycerol was applied directly onto the array and the slide was covered with a 24 × 50 mm glass coverslip (Fisher Scientific, Tustin, CA, USA). A charged coupled device camera was used to image the arrays and intensity data were acquired through DAPI, Cy-3, and Cy-5 channels.
Data Analysis
The SPOT 2.0 software program (available at http://cc.ucsf.edu/jain/public) was used to calculate image data. An average single centered log2 ratio of test intensity over reference intensity for each clone was calculated from triplicate spots on the array. A global threshold of ±0.20 defined gains and losses for all bacterial artificial chromosome clone log2 ratios. Amplifications and homozygous deletions were scored for those clones with log2 intensity ratios greater than 1.0 or less than −0.75, respectively. This threshold roughly corresponds to two standard deviations from the mean, as confirmed in previous experiments using cell lines with known alterations.
Contiguous alterations were expressed as regional or whole arm changes by calculating the median log2ratio for all bacterial artificial chromosome clones mapping to each altered chromosome region. A threshold of ±0.14 was then applied to the median log2 ratio to define ‘regional’ gains or losses relative to the reference sample.
Results
Histopathology and Immunohistochemistry
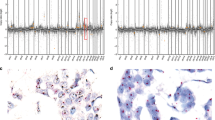
Pathology demonstrated two lesions in the left breast, the known invasive ductal carcinoma (IDC) in the lower inner quadrant, as well as a subcutaneous tumor deposit in the lateral breast. The lower inner quadrant tumor (IDC 1, Figure 1a) was a 1.7-cm, Scarf–Bloom–Richardson grade 2 infiltrating ductal carcinoma. It was associated with extensive, intermediate grade ductal carcinoma in situ. The lateral tumor (IDC 2, Figure 1b) was a 0.8-cm Scarf–Bloom–Richardson grade 2 infiltrating ductal carcinoma. The lesion was located in the superficial breast parenchyma and the invasive tumor was admixed with benign breast lobules and a small component of intermediate grade ductal carcinoma in situ. A sentinel lymph node biopsy showed a 9 mm focus of metastatic adenocarcinoma. Both the breast tumors and the metastatic deposit showed similar histologic features and biologic markers including strong positive staining for estrogen receptor and progesterone receptor and lack of Her-2/neu oncoprotein overexpression. Completion axillary lymph node dissection resulted in retrieval of 17 additional nodes, none of which contained metastatic disease.
Histologic evaluation of the thigh lesion showed a 1.0 cm high-grade leiomyosarcoma with pleomorphic nuclei and small areas of necrosis (Figure 1c). The tumor stained strongly positive for desmin and smooth muscle actin and was negative for S100. Comparison of this lesion to slides from the previously resected retroperitoneal leiomyosarcoma showed similar morphology and immunohistochemical profile (Figure 1d).
Array-Based CGH
Three distinct genetic profiles were observed among these tumors. While the two IDC (cases IDC 1 and IDC 2) were found to be very different, the retroperitoneal and thigh leiomyosarcomas were strikingly similar. Furthermore, the pattern of alterations shared by the leiomyosarcomas differed from the two distinctly different patterns seen in the breast lesions. Both leiomyosarcomas showed many more chromosomal changes than the breast cancers. Within the leiomyosarcomas, 1288 and 921 bacterial artificial chromosome clones were altered in the retroperitoneal and thigh leiomyosarcoma respectively, representing 61% and 43% of the genome. In cases IDC 1 and IDC 2, 280 clones and 554 clones were altered respectively, representing 18% and 24% of the genome.
In the breast lesions, over twice as many clones within case IDC 2 (284 clones (13%)) were gained in comparison to case IDC 1 (127 clones (6%)). Losses were seen in 153 (13%) clones belonging to case IDC 1 and in 270 (12%) clones in case IDC 2. No amplifications or homozygous deletions were observed in either of the two breast lesions.
Most genomic alterations in the leiomyosarcoma tumors (817 (37%) and 735 (34%) clones in retroperitoneal and thigh leiomyosarcomas) involved loss of genomic material when compared to normal reference DNA. Of these clones that were lost, 90 (4%) and 64 (3%) clones were homozygous deletions within the retroperitoneal and thigh leiomyosarcoma. Gains were seen in 186 (8%) and 471 (21%) clones in the retroperitoneal and thigh leiomyosarcomas. In total, 24 (1%) and 13 (<1%) of these clones showed amplifications within the retroperitoneal and thigh leiomyosarcomas respectively.
The pattern of genomic alterations was dissimilar in the two breast cancers (Figure 2). In the first lesion (case IDC 1), five chromosomes exhibited genetic changes. Chromosomes 1 and 3 both have loss in the p arm and gain in q (Table 1). There was a gain in 16p followed by losses of 16q and 17p. Interestingly, loss of chromosome 22, seen in neither the second breast lesion (case IDC 2) nor the leiomyosarcomas, was present as well. In contrast, the second breast lesion (case IDC 2) had a higher number of chromosomal changes and a different pattern of genetic alterations. While the proximal portion of chromosome arm 1p is gained in the second breast lesion (case IDC 2), this region is lost (1p22–1p13, 99.3–133.9 Mb) in the first breast lesion (case IDC 1). Additional changes unique to the second breast lesion (case IDC 2) include low level gains on chromosomes 7, 8, 14, and 18. The only changes found in both breast lesions were 1q gain, 16q loss, and 17p loss. The lack of a shared pattern of changes, with the exception of 1q gain and 16q loss, indicates that the two breast lesions are not clonally related.
Unlike the invasive breast tumors, the genetic profiles of the two leiomyosarcomas were virtually identical. Array CGH of the two leiomyosarcomas are superimposed in Figure 3, with the specific changes shown in Table 2. Both leiomyosarcomas shared loss of either whole chromosomes or of chromosome arms. In addition to these changes, two regions of high level amplifications on 3p23–3p24–3p22 (35.2–37.2 Mb) and 17p12 (9.1–22.6 Mb) were common to both lesions. The two leiomyosarcomas also shared numerous individual clones exhibiting unique gains and losses. The only notable difference was seen in distal chromosome arm 1p, which was partially retained in the thigh leiomyosarcoma but entirely lost in the previously diagnosed retroperitoneal leiomyosarcoma. When array CGH profiles of all four tumors were compared, neither of the leiomyosarcomas was found to have similar changes to either of the invasive breast lesions. The only exception was the common loss of the whole arm 16q.
Discussion
Both DNA and cDNA microarray technology have allowed unprecendented detailed characterization of tumor genome and RNA transcripts. While gene expression profiling has been applied to a wide variety of tumors and has been useful in identifying clinically new subtypes of tumors,6, 7, 8 such analysis has been hampered by the limited availability of frozen tumor specimens. In contrast, array CGH may be performed on archival paraffin-embedded material, although one potential drawback of genomic analysis is that genetic changes alone may not determine phenotype. Both approaches, however, have suffered from lack of standardization in protocol design and data interpretation, impacting the generalizability of new findings.
In this report, we used array CGH to define the genomic relationship between four archived tumors arising in the same patient, three of which were diagnosed synchronously. The almost identical genetic profiles of a previously diagnosed retroperitoneal leiomyosarcoma and the new thigh leiomyosarcoma strongly support that the two lesions were clonally related and that therefore, the thigh lesion represented a metastatic recurrence of the initial primary tumor. In contrast, the individual genetic profiles of the two breast tumors showed distinctly different changes with the exception of 1q gain and 16q loss, suggesting that the two lesions represented two separate foci of multifocal disease rather than a primary cancer with a subcutaneous metastatic deposit.
In previous studies using CGH, leiomyosarcoma has been characterized by numerous losses than gains with the most frequently seen change being loss of 13q as illustrated in the two cases of leiomyosarcoma presented here.9, 10, 11, 12 The chromosome arm of 13q is the location of the RB1 tumor suppressor gene and its deletion may play a role in the tumorigenesis of leiomyosarcoma.10, 13 Additionally, loss of 10q and gain of 17p, both seen in these cases, were also reported previously by CGH.9, 10, 11
Such CGH studies can also contribute to our understanding of malignant progression. While the retroperitoneal leiomyosarcoma and subsequent thigh leiomyosarcoma clearly appear clonally related, we noted the unusual finding of the restoration of distal 1p loss in the metastasis when compared to the primary. This finding may be explained by the possibility that while both tumors shared a common precursor, they then progressed on two separate pathways. The precursor clone at distal chromosome arm 1p (1p36.3) may have been lost during progression to the retroperitoneal leiomyosarcoma whereas it was retained (1p36.3–1p31) in the thigh leiomyosarcoma.
In breast tumors, CGH has been an important research tool for many years, defining the specific genomic characteristics of different tumor types as well as determining tumor progression pathways.14, 15, 16, 17, 18, 19, 20 The cumulative data have contributed to our understanding of the genomic heterogeneity of breast tumors and work is ongoing to determine how genomics determines both tumor phenotype and prognosis. However, there has been no single platform on which this work has been conducted, and in an era of array CGH, lack of array standardization has impeded direct comparison among groups.
For this patient, the CGH findings had important clinical implications. The synchronously diagnosed tumors consisted of two infiltrating ductal carcinomas of the left breast and a leiomyosarcoma of the right thigh; the fourth was a primary left retroperitoneal leiomyosarcoma diagnosed 5 years previously. The clinical relationship between these tumors was unclear; the second breast lesion may have represented a second primary cancer or alternatively a metastatic deposit. The thigh leiomyosarcoma was likely an unusual recurrence of the prior contralateral retroperitoneal leiomyosarcoma, but it may also have been a second primary leiomyosarcoma. CGH analysis was definitive in defining the relationships between these four tumors.
Although the patient had a diagnosis of node-positive invasive breast cancer, the confirmation of Stage IV recurrent leiomyosarcoma by array CGH was consistent with the previous decision to treat the patient with aggressive hormonal therapy only, without chemotherapy. Another clinical scenario in which such CGH data may be of extreme importance are those cases in which a patient develops a metachronous tumor in the breast which harbored a previous breast cancer. Genomic confirmation of either local recurrence or new primary could have profound influence on the treatment recommendations.
We conclude that in cases where multiple synchronous tumors are encountered, genetic analysis by array CGH can clearly elucidate the relationships between different tumor sites, and has the potential to impact clinical treatment decision-making. Especially in those cases where differentiation between metastatic disease and a new primary focus would have important treatment implications and immunohistochemistry is not contributory, array CGH should be considered a potential tool that can offer a definitive answer. However, more widespread clinical use of this technology will necessitate the establishment of standardized methods by which to analyze clinical specimens.
References
Waldman FM, DeVries S, Chew KL, et al. Chromosomal alterations in ductal carcinomas in situ and their in situ recurrences [In Process Citation]. J Natl Cancer Inst 2000;92:313–320.
Gong G, DeVries S, Chew KL, et al. Genetic changes in paired atypical and usual ductal hyperplasia of the breast by comparative genomic hybridization. Clin Cancer Res 2001;7:2410–2414.
Hwang ES, Nyante SJ, Chen YY, et al. Clonality of lobular carcinoma in situ and synchronous invasive lobular cancer. Cancer 2004;100:2562–2572.
Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 1998;20:207–211.
Wilhelm M, Veltman JA, Olshen AB, et al. Array-based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res 2002;62:957–960.
Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–10874.
Linn SC, West RB, Pollack JR, et al. Gene expression patterns and gene copy number changes in dermatofibrosarcoma protuberans. Am J Pathol 2003;163:2383–2395.
Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–511.
Otano-Joos M, Mechtersheimer G, Ohl S, et al. Detection of chromosomal imbalances in leiomyosarcoma by comparative genomic hybridization and interphase cytogenetics. Cytogenet Cell Genet 2000;90:86–92.
Derre J, Lagace R, Nicolas A, et al. Leiomyosarcomas and most malignant fibrous histiocytomas share very similar comparative genomic hybridization imbalances: an analysis of a series of 27 leiomyosarcomas. Lab Invest 2001;81:211–215.
El-Rifai W, Sarlomo-Rikala M, Knuutila S, et al. DNA copy number changes in development and progression in leiomyosarcomas of soft tissues. Am J Pathol 1998;153:985–990.
Parente F, Grosgeorge J, Coindre JM, et al. Comparative genomic hybridization reveals novel chromosome deletions in 90 primary soft tissue tumors. Cancer Genet Cytogenet 1999;115:89–95.
Wang R, Titley JC, Lu YJ, et al. Loss of 13q14–q21 and gain of 5p14–pter in the progression of leiomyosarcoma. Mod Pathol 2003;16:778–785.
Buerger H, Otterbach F, Simon R, et al. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol 1999;189:521–526.
Buerger H, Schmidt H, Beckmann A, et al. Genetic characterisation of invasive breast cancer: a comparison of CGH and PCR based multiplex microsatellite analysis. J Clin Pathol 2001;54:836–840.
Nishizaki T, Chew K, Chu L, et al. Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int J Cancer 1997;74:513–517.
Pollack JR, Perou CM, Alizadeh AA, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 1999;23:41–46.
Thor AD, Eng C, Devries S, et al. Invasive micropapillary carcinoma of the breast is associated with chromosome 8 abnormalities detected by comparative genomic hybridization. Hum Pathol 2002;33:628–631.
Tirkkonen M, Tanner M, Karhu R, et al. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer 1998;21:177–184.
Waldman FM, Hwang ES, Etzell J, et al. Genomic alterations in tubular breast carcinomas. Hum Pathol 2001;32:222–226.
Acknowledgements
We are grateful to the patient who graciously permitted the pathology specimens to be released for CGH analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wa, C., De Vries, S., Chen, Y. et al. Clinical application of array-based comparative genomic hybridization to define the relationship between multiple synchronous tumors. Mod Pathol 18, 591–597 (2005). https://doi.org/10.1038/modpathol.3800332
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800332
Keywords
This article is cited by
-
A rare case of long-term survival of a patient who underwent radical operations for sextuple malignancies
Clinical Journal of Gastroenterology (2022)
-
Distinct genetic evolution patterns of relapsing diffuse large B-cell lymphoma revealed by genome-wide copy number aberration and targeted sequencing analysis
Leukemia (2016)
-
Tiling array-CGH for the assessment of genomic similarities among synchronous unilateral and bilateral invasive breast cancer tumor pairs
BMC Clinical Pathology (2008)
-
Genetic analysis of multifocal superficial urothelial cancers by array-based comparative genomic hybridisation
British Journal of Cancer (2007)
-
Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization
Laboratory Investigation (2006)