Abstract
Human papillomavirus is known to play an important etiological role in the genesis of cervical cancer, but only a very small proportion of infected women develop invasive cervical cancer. The purpose of cervical cancer prevention is early diagnosis of its precursors. The molecular detection of human papillomavirus DNA as a diagnostic test to cervical carcinogenesis gave a low positive predictive value as compared to the use of biomarkers. p16INK4A and possibly p14ARF have been proposed as putative surrogate biomarkers that would allow identification of dysplastic cervical epithelia. Serial consecutive biopsies representing normal cervical epithelium to cervical intraepithelial neoplasia and/or invasive cervical cancer were stained with immunohistochemistry for p16INK4A, p14ARF and proliferating cell nuclear antigen. The positive rates of these markers were significantly higher in cervical intraepithelial neoplasia and in squamous cell carcinoma than in normal cervix (P<0.01). No significant difference was noted between lesions progressing from cervical intraepithelial neoplasia to squamous cell carcinoma for both p16INK4A and p14ARF expression (P>0.05). For both biomarkers, nuclear staining was predominantly seen. However, the cytoplasmic stain of p16INK4A increased with disease progression and the pattern of expression varied between different tumors and its location within the lesion. Both nuclear and cytoplasmic staining with p16INK4A and p14ARF of affected epithelial cells were considered positive. In the adjacent normal tissue to cervical neoplasia, the positive rates of p16INK4A, p14ARF and proliferating cell nuclear antigen expression were higher than those found distant to these lesions but the findings did not reach statistical significance. No correlation was seen between the human papillomavirus types detected and the expression of p16INK4a and p14ARF. In conclusion, overexpression of p16INK4A and p14ARF act as potential biomarkers for cervical cancer progression from premalignant lesions.
Similar content being viewed by others
Main
Cervical cancer is one of the most common cancers affecting women worldwide. Since the implementation of Pap smear screening, cervical cancer morbidity and mortality have declined drastically. Nevertheless, the number of newly diagnosed cases worldwide is still significantly large, reaching about 400 000 cases each year.1 Early diagnosis or trend prediction of cervical lesion development is the main purpose for cervical cancer prevention and treatment. Human papillomavirus (HPV) is known to play an important etiological role in the genesis of cervical cancer. The virus is detected in 95–100% of invasive cervical cancer. High-risk types such as HPV (16, 18, 31, 33, 45, 51) are among most common types found in cervical cancers and are the main factors implicated in cervical carcinogenesis.2, 3
Biologically, cervical cancer arises as a result of an accumulation of damages to cell cycle control and genomic instability. The process of cervical carcinogenesis can take from 5 to 15 years. The number of women infected with high-risk HPV in a population of women aged 20 to 35 years can be as high as 30%. Most infections clear over a period of 10–16 months and of all those infected with HPV only a very small proportion ever develop invasive cervical cancer.4, 5 From the data obtained from epidemiological studies, women with high-risk HPV infections have at least a 100 times higher risk for developing cervical precancer lesions than uninfected women. HPV is recognized and accepted as necessary for cervical carcinogenesis, but the detection of HPV DNA in cervical swabs has a low positive predictive value for cervical neoplasia whereas biomarkers of carcinogenesis have a higher positive predictive value.6, 7 In our recent study, we supported that biomarkers such as p16INK4A could serve as a potential marker for prediction of cervical cancer.8
Several other studies have demonstrated that the p16INK4A protein can be regarded as a specific biomarker that allows identification of dysplastic cervical epithelia with the potential to progress to invasive cervical cancer.9, 10, 11, 12, 13 The tumor suppressor p16INK4A, one of the CDKN2A gene products is a cyclin-dependent kinase inhibitor that negatively regulates cell proliferation by inhibiting hyperphosphorylation of the retinoblastoma tumor suppressor (Rb) by cyclin D-cdk4/cdk6 complexes.14 p16INK4A is involved in the regulation of the cellular life span and the protein accumulates in senescent cells cultured in vitro. Loss of p16INK4A function predisposes mice to tumor development and in particular to carcinogen-induced tumorigenesis.15, 16, 17
p14ARF, also known as p19ARF in mouse, is a putative second surrogate marker for cervical neoplasia following p16INK4A staining.12, 18 p14ARF is encoded in part by a common region in exon 2 shared with p16INK4A but in alternative reading frame. p14ARF is a small basic arginine-rich nuclear/nucleolar protein that can arrest cell growth in G1–S and G2–M in a p53-dependent manner and its expression is increased after oncogenic stress or cell cycle deregulation.19, 20 Thus, p14ARF and p16INK4 are unrelated from structural and functional aspects, although both are tumor suppressors.
Point mutations, promoter hypermethylation and deletions of p16INK4A and p14ARF are frequently found in a variety of human malignancies such as melanoma, lymphoma, lung cancer and bladder cancer.21 In squamous cell carcinoma of the cervix, mutation or homozygous deletions are usually absent, and instead high expression of p16INK4A protein is often observed.13 Presumably, HPV oncoprotein E7 expression leads to inactivation of Rb with subsequent release of the E2F-1 transcription factor that in turn activates p16INK4A and p14ARF transcription.22 p14ARF overexpression may also in part be attributable to functional inactivation of the p53 protein by HPV oncoprotein E6 and subsequent loss of negative feedback control.19
The relation between HPV infection, p16INK4A and p14ARF expression requires further clarification, and in order to evaluate the immunohistochemical staining patterns of these markers we have used cervical biopsies representing different grades of neoplasia as our model.
Expression of p16INK4A, p14ARF and proliferating cell nuclear antigen (PCNA) were detected with immunohistochemistry in 16 cases (38 samples) with serial consecutive biopsies of women who had biopsies representing normal cervical epithelium to cervical intraepithelial neoplasia (CIN) and/or invasive cervical cancer. The aims of this study were (1) to analyze and characterize the expression of p16INK4A, p14ARF and PCNA in cervical lesions, (2) to compare the expression of these proteins between the various histopathological grades of cervical neoplasia, and (3) to evaluate the expression in relation to HPV DNA status.
Materials and methods
Case Selection
Women who developed invasive squamous cell carcinoma who had earlier precancer diagnoses on histology were identified through linkage between the national cancer registry and the pathological registry of Umeå University Hospital. These women were referred for treatment due to an abnormal Pap smear during gynecological examination. To evaluate the potential role of biomarkers, serial consecutive biopsies of these women were selected as they represent the natural history of cervical cancer progression. A total of 19 women who fit the criteria were identified between 1982 and 2000 (for further details see Wang et al8). Two cases were excluded due to insufficient material in their paraffin blocks. The mean age of these women was 38 years of age (range 21–78 years). Each case had two to three consecutive biopsies with diagnosis of normal squamous epithelium, CIN or invasive squamous cell carcinoma. Of these, eight biopsies had only normal epithelium and 11 had only squamous cell carcinoma (see Table 1). The average time interval between the first with a CIN diagnosis to invasive cancer diagnosis was 89.9 months (range: 16–212 months). Overall, 19 biopsies had normal epithelium, CIN and/or squamous cell carcinoma within the same section. The diagnosis of all cases was reviewed and reconfirmed by a pathologist. Owing to problems during sectioning or insufficient material not all paraffin sections of the biopsies were analyzed.
Immunohistochemistry
All specimens were formalin-fixed and paraffin embedded. Serial sections (4 μm thick) were cut and the end section was stained with hematoxylin and eosin to ensure that the lesion was still present in the serial sections. The sections were processed for immunohistochemical staining as previously described.8 The following primary antibodies were used in this study: mouse monoclonal F-12 anti-p16INK4A (Santa Cruz, CA, USA); rabbit polyclonal Ab4 anti-p14ARF (Neomarkers, Fremont, CA, USA); mouse monoclonal PC-10 anti-PCNA (Santa Cruz). Antibodies were diluted 1:100. Secondary antibodies goat anti-mouse IgG diluted in 1:100 (Amersham Biosciences AB, Uppsala, Sweden), donkey anti-rabbit IgG that was used according to instructions in the commercial kit (DAKO K4008). Scoring of immunohistochemistry results were performed on basis of both the staining intensity and distribution of immunopositive cells.12, 23, 24, 25 Staining intensity score: − (minus): no expression, +: weak expression, ++: moderate expression, +++: strong expression. Nuclear staining was considered positive for PCNA, while nuclear staining with or without cytoplasmic staining was considered positive for p16INK4A and p14ARF. The percentage of immunopositive epithelial cells in the total of normal or neoplastic cells of more than 20% was considered positive for p16INK4A, p14ARF and PCNA.12, 23, 24, 25
HPV Detection and Typing
HPV DNA was detected by polymerase chain reaction (PCR). Genomic DNA was extracted from the paraffin-embedded tissue sections. HPV was detected by nested PCR using MY09/11and GP5+/6+ primer sets. They targeted the HPV L1 open reading frame. The PCR products of MY09/MY11 were 450 bp long and GP5+/6+150 bp long. The PCR reaction has been described earlier in Wang et al.8 PCR targeting human S14 gene was used to evaluate the presence of the DNA extractions in these samples. CaSki cell line DNA was used as positive control and blanks were included to check for contamination. HPV typing was obtained from the sequences generated by direct sequencing of purified PCR products using ABI 377 (Applied Biosystems, Foster City, CA, USA). PCR products that could not be sequenced by direct DNA sequencing was sequenced by Pyrosequence.26
Statistical Analysis
SPSS program was used to analyze the data. Statistical analysis was based on χ2-test with Yates' correction. An additional two-tailed Fisher's exact test was used only when the number of samples in any well of a given statistical table was five or fewer. The t-test was used to analyze quantitative data. It was considered significant difference when P-value is less than 0.05.
Results
Expression of p16INK4A in Cervical Neoplastic Lesions
Several studies have reported the predominant expression of p16INK4A in the cytoplasm of cells of metaplasia, dysplasia, high-grade squamous cervical intraepithelial neoplasia and invasive cancer.11, 27 Other investigators have considered only nuclear staining of p16INK4A as positive.28, 29 We used mouse monoclonal antibody F12 to detect p16INK4A protein expression in paraffin-embedded cervical tissues.
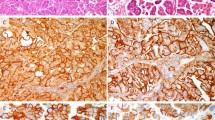
In our material, p16INK4A was expressed predominantly in the nucleus of some cells in normal epithelium, especially those adjacent to cervical lesions. Similar staining pattern was commonly seen in CIN and in some squamous cell carcinoma. Both nuclear and cytoplasmic p16INK4A staining was evident only in approximately 10–20% of invasive cancer cells. Whereas the overall positivity rate for p16INK4A ranges between 10 and 38% of the entire lesion and these cells were mainly found at the basal layer of normal epithelia and in CIN 1 and CIN 2 (Figure 1a, Table 2). As the disease progresses, the staining intensity of the cells and the number of positive cells also increased. In high-grade CIN and invasive cancer, overexpression was more evident as compared to low-grade CIN or normal epithelium. Totally, 21% (4/19) of the high-grade CIN in our material had positively stained cells (nuclear or/and cytoplasm) located from the basal layer up to the lower one-third or two-thirds of the thickness of the entire epithelium, while almost 70% (13/19) of these lesions had positively stained cells throughout the full thickness of the epithelium. Among the invasive cancers, 10/11 cases showed positively stained cells in the entire tumor (Figure 1b and c).
Expression of p14ARF
We detected p14ARF protein only in the nuclei of cells in normal epithelia of the cervix and in low-grade CIN (Figure 1d), while both nuclear/nucleolar and cytoplasmic staining were observed in dysplastic cells of high-grade CIN and invasive cancer (Figure 1e and f). Expression of p14ARF was often seen in the entire CIN lesion where the dysplastic cells were or in the entire tumor. However, some of the invasive cancers stained positively only at the peripheral edge of the tumor towards the stroma. The pattern of expression with p14ARF was rather similar to that of p16INK4A. The difference being in some cases that p14ARF positive cells were more evenly distributed throughout the full thickness of the epithelium in normal, CIN 1 or CIN 2 than the corresponding p16INK4A staining.
Expression Pattern of PCNA in Relation to p14ARF and p16INK4A
PCNA functions as an auxiliary factor for DNA polymerase, and it is involved in DNA repair, as well as in replication. A significant correlation between PCNA staining and the grade of CIN have been found.30 In our material, PCNA staining as detected with PC10 monoclonal antibody was mainly nuclear in normal epithelium, CIN or squamous cell carcinoma (Figure 1g–i). Of the 19 CINs, PCNA-positive cells that located at the upper one-third of the cervical epithelium were seen in two samples (11%), or located at the lower one-third to half of the thickness of the epithelium in five samples (26%). In nine samples (53%) the distribution of positive cells were seen in the entire epithelium. Interestingly, in invasive cancer, PCNA-positive cells were distributed at the edge of the lesion occupying one-third of the tumor in three samples, whereas in the other samples/sections, PCNA-positive cells were seen in the entire cancerous lesion (8/11).
p16INK4A, p14ARF and PCNA Expression Rates in CIN and Invasive Cervical Cancer
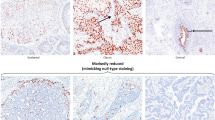
The positive rates of p16INK4A, p14ARF and PCNA expression in normal cervix, CIN and squamous cell carcinoma are shown in Table 1. Overall, the positive rates of these three biomarkers were significantly higher in CIN and in squamous cell carcinoma than in normal cervix (P<0.01). However, no significant difference was noted between lesions progressing from high-grade CIN to squamous cell carcinoma for p16INK4A and p14ARF expression (P>0.05). Notably, the positive rate of PCNA in squamous cell carcinoma (100%) was significantly higher than in CIN (60%) (P<0.05). The expression of p16INK4A, p14ARF and PCNA between normal epithelia alone and normal epithelia adjacent to CIN or squamous cell carcinoma was also compared. Normal epithelium adjacent to CIN had more positive cells for these biomarkers (Figure 2) than normal epithelium not adjacent to any CIN or squamous cell carcinoma. The positive rates of p16INK4A, p14ARF and PCNA in the adjacent normal tissue were 38, 31 and 13%, respectively, which were higher than that of normal epithelia alone (p16INK4A 13%, p14ARF 25% and PCNA 0%, see Table 1). However, these differences were not statistically significant (P>0.05).
p16INK4A and p14ARF Expression in Relation to HPV Infection
The data on p16INK4A, p14ARF and PCNA expression and HPV status are shown in Table 2. In all, 17 (90%) cases were positive for HPV DNA. The main HPV types were HPV 16 (41%), HPV33 (24%), HPV18 (18%) and HPV45 (12%). One sample was HPV DNA positive but could not be typed according to DNA sequence. HPV DNA-positive rate was 63% in normal tissue, 90% in CIN and 100% in squamous cell carcinoma. Among the 17 HPV DNA positive CINs, the positive rates of p16INK4A, p14ARF and PCNA expression were 77 (13/17), 77 (13/17) and 88% (15/17), respectively. In the two HPV DNA-negative cases 1a and 4a, p16INK4A was positive in both cases, while p14ARF and PCNA were positive only in case 4a. No direct correlation between the individual HPV types detected in each tissue section and the expression rates of each of these biomarkers was noted.
Discussion
High-risk HPV infection is critically involved in cervical carcinogenesis though not all infections led to the development of invasive cervical cancers. High-risk HPV encoded proteins play an important role in the control and inactivation of the Rb and p53 tumor suppressors: of the cell cycle regulatory cascade through the binding of its oncoproteins E6 and E7 to the tumor suppressor genes p53 and Rb.31 Cervical cancers are most often HPV related and therefore the p16INK4A and p14ARF status of cervical cancers could differ from non-HPV-related cancers of other anatomical sites. Interference or abrogation of the p53 and Rb cell cycle pathways may cause the induction of p16INK4A and p14ARF transcription. Therefore, most high-risk HPV-related neoplasia stained positively for these proteins by immunohistochemistry.
Earlier studies have shown a predominant of cytoplasmic p16INK4A staining using immunohistochemistry.11 Murphy et al9 found a small number of CIN 1 lesions exhibiting exclusively nuclear staining, whereas CIN 2 and CIN 3 lesions had a predominantly cytoplasmic staining pattern while all the invasive squamous cancers exhibited strong nuclear and cytoplasmic staining. Hence, either strong nuclear or cytoplasmic staining had been considered positive for p16INK4A. The results obtained in our study showed p16INK4A expression as predominantly in the nuclei of most of our tumor samples, while only a few of these tumors displayed both nuclear and cytoplasmic staining. Thus, the overexpression of these biomarkers in both nuclear and cytoplasm of affected cells are to be considered positive.
p14ARF, the alternative reading frame product of CDKN2A, activates a p53 response that manifests itself as elevated levels of MDM2, p21, and cell cycle arrest in both G1 and G2/M phases.19, 20 Several studies have detected p14ARF expression with immunohistochemistry using monoclonal and polyclonal anti-p14ARF antibodies in the nucleus/nucleolus of primary tumors such as cervical cancer, nasopharyngeal carcinomas, non-Hodgkin's lymphoma and in various brain tumors.12, 18, 28, 32, 33, 34 In our material, p14ARF staining was predominantly nuclear. Although, p14ARF presents it as a nucleolar protein in human tumor cell lines,35 localization of the protein has also been detected in the nucleoplasm as reported elsewhere and in some cases36 it is associated with aggressive growth and a malignant phenotype.34 Sano et al12, 18 described different types of p14ARF expression patterns: those staining the top half layer of the epithelium (the superficial type) and those staining the full thickness type. p14ARF staining was especially prominent in cells showing ‘koilocytotic change’ or in cells with marked nuclear pleomorphism,12 while we detected p14ARF-positive cells in the entire CIN and invasive cancer lesion.
p16INK4A and p14ARF are both strongly expressed in high-grade lesions and in invasive cervical cancer but not significantly in normal squamous epithelium of the cervix.9, 11, 12, 37 Thus, these proteins are considered potential biomarkers for abnormal squamous epithelial dysplasia and invasive cancer. Our study supports previous findings in that both p16INK4A and p14ARF expression in the high-grade CIN and squamous cell carcinoma of our progressing cases were significantly higher than the normal cervical epithelium present simultaneously within the same tissue sections (P<0.01). However, the expression rate between the high-grade CIN and squamous cell carcinoma was not statistically significant (P>0.05).
PCNA is an S-phase-associated nuclear protein. Monoclonal antibodies to PCNA can be used as an S-phase probe on paraffin-embedded tissue.38 PCNA staining is often evident in basal as well as suprabasal layers of cervical dysplasia, whose expression corresponded to and clearly declined with the grade of squamous intraepithelial lesions. The relatively long half-life of PCNA coupled with rapid cell turnover resulted in such seemingly aberrant suprabasal staining.39 In our study, PCNA expressing cells were located in the upper one-third in two cases or the lower one-third to one-half of the epithelium of CIN lesions in five cases. The positive rate of PCNA expression was significantly increased during progression from normal to CIN and was also increased from CIN to squamous cell carcinoma.
In this study, the 17 cases were HPV DNA positive, and the approximate percentage positive rates for p16INK4A, p14ARF and PCNA overexpression were rather similar (77–88%). Theoretically, all high-risk HPV-associated lesions should express p16INK4A and p14ARF. However it is unclear why 29% (5/17) of our samples that were HPV DNA positive were also p16INK4A negative regardless of the histological diagnosis of the lesions. We might assume that there are other mechanisms of p16INK4A regulation besides HPV infection, such as promoter methylation, that could occur in cervical cancer. We noted a slight increase in expression of p16INK4A and p14ARF in normal epithelium adjacent to CIN or invasive cervical cancers. This indicates abnormalities in normal cell functions in these cells despite their apparent normal morphology. Presumably, the microenvironment of the adjacent ‘normal’ epithelia is different to that distant to any dysplastic lesions. In conclusion, we confirm in this study the presence of p16INK4A and p14ARF expression as potential biomarkers for cervical cancer progression from premalignant lesions.
References
Parkin DM . Global cancer statistics in the year 2000. Lancet Oncol 2001;2:533–543.
van der Graaf Y, Molijn A, Doornewaard H, et al. Human papillomavirus and the long-term risk of cervical neoplasia. Am J Epidemiol 2002;156:158–164.
Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–19.
Hildesheim A, Schiffman MH, Gravitt PE, et al. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis 1994;169:235–240.
Ho GY, Bierman R, Beardsley L, et al. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med 1998;338:423–428.
von Knebel Doeberitz M . New molecular tools for efficient screening of cervical cancer. Dis Markers 2001;17:123–128.
von Knebel Doeberitz M . New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer 2002;38:2229–2242.
Wang JL, Zheng BY, Li XD, et al. Predictive significance of the alterations of p16INK4A, p14ARF, p53, and proliferating cell nuclear antigen expression in the progression of cervical cancer. Clin Cancer Res 2004;10:2407–2414.
Murphy N, Ring M, Killalea AG, et al. p16INK4A as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep smears. J Clin Pathol 2003;56:56–63.
Klaes R, Benner A, Friedrich T, et al. p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol 2002;26:1389–1399.
Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 2001;92:276–284.
Sano T, Masuda N, Oyama T, et al. Overexpression of p16 and p14ARF is associated with human papillomavirus infection in cervical squamous cell carcinoma and dysplasia. Pathol Int 2002;52:375–383.
Sano T, Oyama T, Kashiwabara K, et al. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol 1998;153:1741–1748.
Serrano M . The tumor suppressor protein p16INK4a. Exp Cell Res 1997;237:7–13.
Hara E, Smith R, Parry D, et al. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol 1996;16:859–867.
Krimpenfort P, Quon KC, Mooi WJ, et al. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 2001;413:83–86.
Sharpless NE, Bardeesy N, Lee KH, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 2001;413:86–91.
Sano T, Hikino T, Xue Q, et al. Immunohistochemical inactivation of p14ARF concomitant with MDM2 overexpression inversely correlates with p53 overexpression in oral squamous cell carcinoma. Pathol Int 2000;50:709–716.
Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 1998;17:5001–5014.
Sherr CJ, Weber JD . The ARF/p53 pathway. Curr Opin Genet Dev 2000;10:94–99.
Ruas M, Peters G . The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta 1998;1378:F115–F177.
Dimri GP, Itahana K, Acosta M, et al. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol 2000;20:273–285.
Herbsleb M, Knudsen UB, Orntoft TF, et al. Telomerase activity, MIB-1, PCNA, HPV 16 and p53 as diagnostic markers for cervical intraepithelial neoplasia. APMIS 2001;109:607–617.
Tjalma WA, Weyler JJ, Bogers JJ, et al. The importance of biological factors (bcl-2, bax, p53, PCNA, MI, HPV and angiogenesis) in invasive cervical cancer. Eur J Obstet Gynecol Reprod Biol 2001;97:223–230.
Wong SC, Chan JK, Lee KC, et al. Differential expression of p16/p21/p27 and cyclin D1/D3, and their relationships to cell proliferation, apoptosis, and tumour progression in invasive ductal carcinoma of the breast. J Pathol 2001;194:35–42.
Gharizadeh B, Ghaderi M, Donnelly D, et al. Multiple-primer DNA sequencing method. Electrophoresis 2003;24:1145–1151.
Keating JT, Cviko A, Riethdorf S, et al. Ki-67, cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol 2001;25:884–891.
Korshunov A, Shishkina L, Golanov A . Immunohistochemical analysis of p16INK4a, p14ARF, p18INK4c, p21CIP1, p27KIP1 and p73 expression in 271 meningiomas correlation with tumor grade and clinical outcome. Int J Cancer 2003;104:728–734.
Makitie AA, MacMillan C, Ho J, et al. Loss of p16 expression has prognostic significance in human nasopharyngeal carcinoma. Clin Cancer Res 2003;9:2177–2184.
Shurbaji MS, Brooks SK, Thurmond TS . Proliferating cell nuclear antigen immunoreactivity in cervical intraepithelial neoplasia and benign cervical epithelium. Am J Clin Pathol 1993;100:22–26.
Galloway DA, McDougall JK . The disruption of cell cycle checkpoints by papillomavirus oncoproteins contributes to anogenital neoplasia. Semin Cancer Biol 1996;7:309–315.
Korshunov A, Golanov A, Timirgaz V . p14ARF protein (FL-132) immunoreactivity in intracranial ependymomas and its prognostic significance: an analysis of 103 cases. Acta Neuropathol (Berl) 2001;102:271–277.
Crook T, Nicholls JM, Brooks L, et al. High level expression of deltaN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma. Oncogene 2000;19:3439–3444.
Sanchez-Aguilera A, Sanchez-Beato M, Garcia JF, et al. p14(ARF) nuclear overexpression in aggressive B-cell lymphomas is a sensor of malfunction of the common tumor suppressor pathways. Blood 2002;99:1411–1418.
Lindstrom MS, Klangby U, Inoue R, et al. Immunolocalization of human p14(ARF) to the granular component of the interphase nucleolus. Exp Cell Res 2000;256:400–410.
Geradts J, Wilentz RE, Roberts H . Immunohistochemical [corrected] detection of the alternate INK4a-encoded tumor suppressor protein p14(ARF) in archival human cancers and cell lines using commercial antibodies: correlation with p16(INK4a) expression. Mod Pathol 2001;14:1162–1168.
Sano T, Oyama T, Kashiwabara K, et al. Immunohistochemical overexpression of p16 protein associated with intact retinoblastoma protein expression in cervical cancer and cervical intraepithelial neoplasia. Pathol Int 1998;48:580–585.
Garcia RL, Coltrera MD, Gown AM . Analysis of proliferative grade using anti-PCNA/cyclin monoclonal antibodies in fixed, embedded tissues. Comparison with flow cytometric analysis. Am J Pathol 1989;134:733–739.
Keating JT, Ince T, Crum CP . Surrogate biomarkers of HPV infection in cervical neoplasia screening and diagnosis. Adv Anat Pathol 2001;8:83–92.
Acknowledgements
The study has been supported by grants from the Swedish Cancer Society, the Swedish Cancer foundation, the Swedish Medical Research Council in Stockholm, Sweden and Stockholm County Council.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wang, JL., Zheng, BY., Li, XD. et al. p16INK4A and p14ARF expression pattern by immunohistochemistry in human papillomavirus-related cervical neoplasia. Mod Pathol 18, 629–637 (2005). https://doi.org/10.1038/modpathol.3800308
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800308
Keywords
This article is cited by
-
p16INK4A expression is frequently increased in periorbital and ocular squamous lesions
Diagnostic Pathology (2015)
-
Human Papilloma Virus Early Proteins E6 (HPV16/18-E6) and the Cell Cycle Marker P16 (INK4a) are Useful Prognostic Markers in Uterine Cervical Carcinomas in Qassim Region- Saudi Arabia
Pathology & Oncology Research (2015)
-
p16INK4a and p14ARF mRNA expression in Pap smears is age-related
Modern Pathology (2012)
-
p16 expression is not associated with human papillomavirus in urinary bladder squamous cell carcinoma
Modern Pathology (2012)
-
Molecular Basis for Advances in Cervical Screening
Molecular Diagnosis (2005)