Abstract
Adenoid cystic carcinoma, a relatively uncommon tumor of salivary glands, is characterized by a prolonged clinical course and a fatal outcome. The molecular events underlying their progression are unknown. In this study, we examined the methylation status of E-cadherin gene and its protein expression in 23 cases of adenoid cystic carcinoma and correlated the results with the clinicopathologic factors to determine its role in these tumors. We also analyzed the effect of 5-azacytidine on the re-expression in a methylated cell line of adenoid cystic carcinoma for this gene. In our study, E-cadherin immunoreactivity, although heterogeneous, showed a progressive reduction with high histological grade and in metastatic and recurrent lesions. Promoter methylation was detected in 16 of 23 cases (70%), but there was no correlation with the histological grade or patient prognosis. Microdissection of immuno-negative cells in heterogeneous tumors showed positive methlyation. In the cell line from salivary adenoid cystic carcinoma with methylated E-cadherin, 5-azacytidine restored the E-cadherin expression. Our results indicate that: (1) E-cadherin gene promoter is frequently methylated in adenoid cystic carcinoma, leading to reduced E-cadherin expression, (2) variable E-cadherin expression might result from the intratumoral heterogeneity, and (3) increased extent of methylated areas may be associated with progression and advancement of the disease.
Similar content being viewed by others
Main
Adenoid cystic carcinoma is a relatively uncommon salivary gland tumor characterized by indolent and ultimately fatal course.1, 2, 3 Local and distant metastases are common features of this entity, regardless of the histological features or the status of surgical margins.4, 5 Alterations of cell adhesion molecules or production of excessive matrix protein have been proposed as factors associated with the aggressive behavior in adenoid cystic carcinoma.6, 7 However, the underlying events for the aggressive nature and protracted course of this tumor are poorly understood.
E-cadherin (E-cad), a family of calcium-dependent cell adhesion molecules, possesses intercellular attachment, cell polarity, and morphogenesis functions through the formation of complexes with α- and β-catenin and plakoglobin at the transmembrane domain.8 Abnormal E-cad expressions, including cytoplasmic translocation, heterogeneous, and/or absence of expression have been reported in aggressive carcinomas of the esophagus, stomach, and breast.9, 10 Mutation of the E-cad gene and loss of heterozygosity (LOH) of this gene have also been detected in cell lines of gastric11, 12, 13, 14, 15 and breast carcinomas.16 Previous studies of this gene in adenoid cystic carcinoma have reported a reduced expression of E-cad protein in high grade and aggressive tumors.17 The factors leading to the reduction of expression in adenoid cystic carcinoma, however, remain unknown.
Methylation has recently been shown to be an important mechanism in the transcriptional inactivation of certain tumor suppressor genes.18, 19, 20 We previously reported that promoter methylation of p16 gene, a cell cycle regulator of G1/S boundary, occurs in 20% of adenoid cystic carcinomas and induces the reduction of its protein expression21 Furthermore, methylation of the promoter CpG island of E-cad has been found to accompany abnormal expression of immunoreactive E-cad in gastric, liver, thyroid, breast, and prostate carcinomas.22, 23, 24, 25, 26 These lines of evidence led us to hypothesize that altered E-cad expression by CpG island methylation may underlie the aggressive behavior of adenoid cystic carcinoma during its protracted course.
In the present study, we therefore investigated the methylation status of the E-cad gene and its protein expression in cases with adenoid cystic carcinoma. Laser capture microdissection methods (LCM) were applied to correlate methylation to gene expression in heterogeneous tumors. We further analyzed whether demethylation by 5-azacytidine affects the E-cad expression in cultured cells derived from salivary gland adenoid cystic carcinoma.
Materials and methods
Tissue Samples of Surgical Cases and Cell Culture
In all, 26 lesions from 23 patients with the diagnosis of adenoid cystic carcinoma (23 primary, two recurrent, and one metastatic tumors) were retrieved from the Department of Pathology, Hirosaki University and associated hospitals comprised the materials of this study. Immunohistochemical staining of E-cad protein and DNA extraction for the methylation analysis of the E-cad promoter region were performed on sections from selected paraffin blocks (Table 1). Locations of the primary tumors were: submandibular gland in five cases, parotid gland in four, nasal cavity in three, pharynx in three, palate in two, maxillary sinus in two, lip, tongue, larynx, and external auditory canal, one case at each site, respectively. Two of the recurrent tumors were at the same site as the original tumor and one was from a metastatic lesion in the cranium (Table 1). The clinical and pathological data on the patients were obtained from medical records and surgical pathology reports. All tumors had at least some foci with a cribriform pattern having a variable mixture of tubular and solid areas. In each tumor, we determined the predominant histological pattern and histological grade of malignancy according to the criteria of Szanto et al:27 Grade 1, tumor with tubular and cribriform pattern but without solid component; Grade 2, tumor with a pure cribriform pattern or mixed with less than 30% of solid areas; and Grade 3, tumor with more than 30% of solid areas. Control specimens were obtained from cases of pleomorphic adenoma and Warthin's tumor as well as normal salivary glands.
For the in vitro experiments, the ACC3 cell line, originally derived from a patient of adenoid cystic carcinoma arising from the parotid gland, was used.28 Cultured tumor cells were maintained in RPMI-1640 medium (Gibco BRL, Rockville, MD, USA) supplemented with 10% fetal bovine serum (Gibco BRL), 1% L-glutamine, 50 μg/ml streptomycin, and 50 IU/ml penicillin, and incubated at 37°C under a humidified atmosphere containing 5% CO2.
Immunohistochemistry
Immunohistochemical staining for E-cad was performed on 10% formalin-fixed deparaffinized sections using the streptavidin–biotin method. Antigen retrieval using microwave irradiation in 0.01 mol/l citrate buffer (pH 6.0) was used and the sections were immersed in methanol with 0.3% hydrogen peroxide for 30 min and treated with 10% normal rabbit serum for 30 min for inhibition of endogenous peroxidase activity. The slides were first incubated with anti-human E-cadherin monoclonal antibody (HECD-1, 1:500 dilution; Takara Shuzo, Ohtsu, Japan) overnight at 4°C. They were then incubated at room temperature for 30 min each with secondary biotinylated anti-mouse immunoglobulin antibody and peroxidase-conjugated streptavidin (Histofine SAB kit, Nichirei, Tokyo, Japan). Reaction products were detected with 3,3′-diaminobenzidine tetrahydrochloride. The nuclei were counterstained with hematoxylin. Sections of pleomorphic adenoma and Warthin's tumor were used as positive controls. Negative controls were obtained by staining protocols omitting the first antibody, or by using nonimmune mouse sera in place of the first antibody.
Cells with clear intercellular and contiguous membranous stainings were scored as positive cells for E-cad expression. The number of positive cells in 1000 tumor cells within 4–6 microscopic fields at × 200 magnification was counted. Staining results were classified into four grades depending on the percentage of E-cad-positive cells negative (no positive cells), low (<15% positive tumor cells), moderate (15–50% positive tumor cells), and diffuse (>50% positive tumor cells). Two independent pathologists conducted quantitation of positive cells, and the interobserver concordance was more than 90%.
DNA Extraction
From paraffin-embedded tumor tissues, genomic DNA was extracted from five to 10 serial sections of 10 μm thickness after deparaffinization, based on standardized methods.29 All samples were tested for the presence of β-globin DNA by the polymerase chain reaction (PCR) to determine the sample quality before they were analyzed. Similarly, genomic DNA from ACC3 cells was extracted by standard proteinase K and phenol/chloroform extraction procedures.
Methylation-Specific PCR (MSP)
DNA methylation patterns in the E-cad promoter region were determined by MSP. Methylation of the E-cad promoter gene at the site of the CpG island was examined by MSP using the CpGenome™ DNA Modification Kit (Intergen, Purchase, NY, USA). Extracted DNA samples were bisulfite modified, following the manufacturer's instructions. Bisulfite-modified DNAs were then PCR-amplified using primer pairs of E-cad-U and E-cad-M, as follows:

The construct of the primers contained six sites (116 bp) of CG promoter at genomic position of −205 for E-cad-M and five sites (97 bp) at genomic position of −201 for E-cad–U, respectively.30 The PCR assay was performed in a final volume of 25 μl containing template DNA, 1 × PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2), 0.2 mM of each dNTP, 500 nM of each primer, and 1 U of AmpliTaq gold (Perkin-Elmer, Roche Molecular Systems, Branchburg, NJ, USA). Hot start PCR was used for this assay. PCR was performed at 95°C for 12 min, followed by 35 cycles at 95°C for 1 min, 53°C for 1 min, and 72°C for 2 min, with a final 7 min extension at 72°C. The PCR products were electrophoresed on 2% polyacrylamide gels. Gels were stained with ethidium bromide and visualized by ultraviolet illumination.
PCR products from four cases were sequenced with an Automated DNA Sequencer ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems, Tokyo, Japan).
Laser Capture Microdissection (LCM) Analysis
To examine whether there is a topographic difference between protein expression and methylation status, LCM (PixCell 100 apparatus, Arcturus Engineering Inc., Mountain View, CA, USA) was used to obtain DNA from deparaffinized immunostained sections of E-cad-positive or negative areas. Only cases positive for methylation by MSP were used (cases 5, 13, 15, 22, and 23b).
In vitro Effects of 5-Azacytidine on ACC3 Cells
ACC3 cell line was cultured for 7 days in the presence of the demethylating agent, 5-azacytidine (5AC), and analyzed for the expression of the E-cad gene. Protein and mRNA expressions were also compared with those from untreated cultured cells. ACC3 cells were seeded at a concentration of 1 × 105 cells in a 60 mm plastic dish in medium containing 10 μM 5AC (Sigma, St Louis, MO, USA). The medium was replaced every 2 days by fresh medium. As controls, ACC3 cells were incubated for 7 days with medium without 5AC. The experiments were repeated three times and, in each experiment, at least three dishes were prepared for both 5AC-treated and untreated groups.
The methylation status of the extracted DNA was examined by MSP as described above. Immunostaining was applied to the cultured cells on the coverslip fixed with acetone. Staining procedures were the same as the method applied to the tissue sections. The expression of E-cad mRNA was further evaluated by reverse transcriptase PCR (RT-PCR).
RT–PCR
Total RNA was extracted from cultured ACC cells with or without 5AC treatment by Trizol reagent (Gibco BRL) following the manufacturer's protocol. Normal gastric mucosal tissue was used for comparison. cDNA was prepared by RT of 1 μg of total RNA using SUPERSCRIPT™ first-strand synthesis system (Gibco BRL). PCR was performed in a total volume of 50 μl containing 1 × PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2), 0.2 mM of each dNTP, 200 nM of each primer, 1 U of AmpliTaq gold (Roche Molecular Systems, Branchburg, NJ, USA). The primers used for E-cad were:

as previously described.31 The amplification condition was 94°C for 10 min followed by 40 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final 7 min extension at 72°C. The PCR products were electrophoresed on 4–20% gradient polyacrylamide gels. Gels were stained with ethidium bromide and visualized by ultraviolet illumination.
Statistical Analysis
All statistical evaluations were performed using Stat View 5.0 software (SAS Institute, Cary, CA, USA). The association between histological grade, immunohistochemical expression, and methylation status was assessed by Fisher's exact test and regression analysis. Multivariate analysis was performed using the Cox proportional hazard model to evaluate the relationship between overall survival and clinicopathologic factors. Survival curves were plotted by the Kaplan–Meier method to assess the correlations between each factor and overall survival. Differences in survival curves were estimated by the log-rank test. P-values less than 0.05 were considered significant.
Results
E-cad Expression in Surgical Cases of Adenoid Cystic Carcinoma
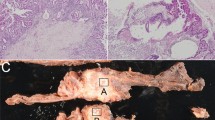
Diffuse membranous staining of E-cad was detected in the ductal epithelial cell components of normal salivary glands. The expression of E-cad in adenoid cystic carcinoma was variable within and between cases. In a tubular growth pattern, E-cad immunoreactivity was homogenously located at the cell surface (Figure 1a). In cribriform and solid growth areas, a strong positive expression was noted in some but negative in others within the same tumors, whereas positive reactions were variable in areas with cribriform growth pattern, structurally consistent with histological Grade 2 (Figure 1b). In areas with a solid growth pattern, histologically consistent with Grade 3, the immunohistochemical reaction was mostly negative (Figure 1c). A completely negative reaction was found in six cases (24%). In particular, in the recurrent and metastatic lesions, expression of E-cad was negative or low, compared with the reactions in the original tumor (Figure 2a, b). The histological grade was weakly correlated with E-cad expression (r2=0.264, P<0.01) (Figure 3).
Expression of immunoreactive E-cadherin in adenoid cystic carcinoma. (a) Diffuse membranous staining was apparent in most tumor cells with tubular growth pattern, consistent with histological Grade 1. (b) In areas of cribriform pattern, consistent with histological Grade 2, expression of E-cadherin was less apparent showing scattered positive cells with membranous or cytoplasmic reactions. (c) There was hardly any positive reaction in solid areas of the tumor, showing invasive growth into the stroma, when histological grade was assigned to Grade 3.
Temporal changes of E-cadherin expression with advancement of disease in adenoid cystic carcinoma (Case 23). The original tumor consisted mostly of tubular growth, showing strong expression of E-cadherin (a), while the growth pattern became solid lacking apparent expression of E-cadherin in the metastatic site of the bone (b).
Promoter Methylation of the E-cad Gene
Methylation of E-cad promoter was detected in 16 of 23 (70%) of the cases (Figure 4) (Table 1). Five of seven cases (71%) of Grade 1 tumors, seven of 11 Grade 2 (64%), and four of five (80%) Grade 3 tumors were positive for methylation, showing no correlation between the incidence of methylation and histological grading (Table 2). However, two cases in which the original tumors were negative for methylation were positive in the recurrent or metastatic lesions (Cases 4 and 23). The latter was originally unmethylated (E-cad expression 80%), and then became methylated in the high-grade tumor at metastatic site (E-cad expression 8%). In all, 10 of 16 (63%) methylation-positive cases were negative or showed low immunoreactivity for E-cad expression (<15% of positive tumor cells), and in contrast, two of seven unmethylated cases were negative or low for E-cad expression (Table 2).
Analysis by methylation-specific polymerase chain reaction on E-cadherin promoter in cases of adenoid cystic carcinoma. All cases presented (cases 8, 10, 16, and 23 b), except for case 23a which showed methylated genes. Normal salivary gland and benign salivary tumors (pleomorphic adenoma and Warthin tumor) were all negative for methylation. U=unmethylated DNA, M=methylated DNA.
Pleomorphic adenomas and Warthin's tumors were negative. E-cad-U and E-cad-M amplification products from cases 11 and 19 were sequenced to confirm the presence of methylation. Conversion from C to T at the CpG site was demonstrated in the sequence of E-cad-U products by bisulfite treatment (Figure 5). There was no alteration in the E-cad-M sequence.
Direct sequencing of the amplified products with E-cad-M (a) and E-cad-U (b) primers in case 19. Base of cytosine (c) in the unmethylated DNA was all converted to thymidine (T) after bisulfide treatment, whereas C in methylated DNA remained as C, indicating the presence of methylation at the CpG sites.
Survival Analysis
Within the follow-up period, 14 of 21 (67%) patients died of cancer recurrence and distant metastasis. The Cox hazard model using six variables (Table 3) revealed that the histological grade was an independent factor that affected the prognosis at the time of initial diagnosis. Similarly, univariate analysis demonstrated that histologically high-grade tumors (Grade 3) showed a significantly shorter survival than other low-grade tumors (Grade 1 and 2) (Figure 6a). On the other hand, other factors such as age, sex, location, E-cad protein expression, or methylation status of E-cad gene did not have a significant influence on the prognosis (Figure 6b).
The relationships between overall survival rate and histologic grade or E-cadherin expression in patients with adenoid cystic carcinoma. (a) Overall survival of the patients with Grade 3 tumors was significantly shorter than that of other grades of tumor. (b) The expression of E-cadherin does not significantly influence the overall survival of the patients.
Microdissection Analysis
The high E-cad expression areas were negative for methylation in all five cases examined, while E-cad negative expression areas were positive for methylation (Figure 7).
Site-specific changes of E-cadherin expression and methylation status in adenoid cystic carcinoma. With the laser capture microdissection method, E-cadherin positive and negative areas were separately examined for the presence of methylation. Tissues that were strongly positive for E-cadherin expression (a) before dissection; ((b) after dissection) were negative for methylation in case 23a, whereas areas of negative for E-cadherin expression (c) before dissection; ((d) after dissection) were positive for methylation.
Methylation and Expressions of E-cad mRNA and Protein in Cultured ACC3 Cell Line
Both unmethylated and methylated bands were detected by MSP on the genomic DNA extracted from ACC3 cell line in the absence of 5AC (Figure 8a). No methylated band was noted in the 5AC-treated cell line (Figure 8b). Immunohistochemical examination showed clear expression of E-cad proteins in cultured ACC cells treated with 5AC. E-cad mRNA expression was equivocal in untreated ACC3 cells, whereas treated cells exhibited clear expression of E-cad mRNA (Figure 8c).
Promoter methylation of E-cadherin gene and its protein expression in cultured ACC3 cells and effects of 5-azacytidine (5AC). Immunohistochemistry demonstrated clear cytoplasmic expression of E-cadherin in 5AC-treated cells (a), while the reactions were completely negative in untreated cells (b). Untreated ACC3 cells showed methylated band of E-cadherin promoter gene in the extracted genomic DNA ((b) inset), whereas there was no methylated band in 5AC-treated cells ((a) inset). The mRNA expression of E-cadherin was also induced after treatment with 5AC in ACC cells by RT–PCR (c).
Discussion
Our study firstly investigated the relationship of methylation of the E-cad gene promoter and its protein expression in adenoid cystic carcinoma. We observed an association between methylation of this gene and the reduced E-cad expression. In situ LCM analysis revealed that areas with high expression were negative for methlyation, whereas E-cad-negative areas were positive for methylation in all the five cases examined. The results of in vitro experiments confirmed that the demethylation status correlated with the expression of E-cad proteins and mRNA transcripts. Interestingly, two cases with recurrent and metastatic lesions, where the original tumors showed a high expression of E-cad without methylation progressed to show methylated genes with a marked reduction of protein expression, indicating an association of methylated status with tumor progression. These findings are in keeping with those of previous studies showing low protein expression in high-grade tumors (Grade 3).17 This further supports the recent proposal that epigenetic changes of the E-cad gene may be a major cause of invasion and metastasis of breast cancer.32 It is of note, however, from our survival analysis that only histological grade at the time of diagnosis predicted the patient prognosis but the correlation between E-cad expression and histological grade was only marginal (r2=0.264, P<0.01). There was no significant correlation between overall methylation status and E-cad expression or prognosis, indicating that the methylation status per se as an all-or-nothing basis does not directly confer the histological grade or predict the prognosis. The extent of methylated areas within a single tumor may rather be relevant to the histological grade or patient prognosis. In addition, we noted reduced expression without methylation in a few of our cases. This may suggest that mechanisms other than methylation are likely to be involved in impaired E-cad expression. Otherwise, restricted use of formalin-fixed paraffin materials as a source of DNA may also lead to false-negative results due to possible marked DNA fragmentations.
Methylation was highly found in cases with different histological grades of adenoid cystic carcinoma (Grade 1: 71%, Grade 2: 64%, and Grade 3: 80%). A high incidence of methylation of the E-cad promoter gene has also been detected in various types of carcinomas.22, 24, 32, 33, 34 Moreover, methylated CpG islands of the E-cad gene were found to be frequent in non-neoplastic bladder mucosa contiguous to cancer lesions.35 However, methylation detections from paraffin sections by MSP should carefully be interpreted in the context of intratumoral heterogeneity and field effect. We observed a discrepancy between immunoexpressions and the methylation status in some of these tumors. Similar findings have been reported in breast cancer, where a striking heterogeneity in methylation between tumors, even in the carefully microdissected in situ, was observed.26 Intratumoral E-cad expression heterogeneity was also noted in other carcinomas.9 In the current study, methylation of E-cad gene was frequent even in low-grade tumors (Grade 1). MSP is a remarkably sensitive method that can detect 0.1% methylated cells in a specimen,30 implicating the possibility that low-grade tumors mostly consisted of E-cad protein positive cells with only a very minor proportion of methylated cells. This assumption may be supported by our findings from in situ LCM analysis, which demonstrated the association between the protein expression and the methylation status in the heterogeneous components within a single tumor.
Our MSP analysis of the ACC3 cell line showed that E-cad gene in the genomic DNA extracted from this cell line was highly methylated and this can be inhibited after 5AC-treatment. In addition, the effect of 5AC-treatment clearly reflected the expression of E-cad proteins and mRNA transcripts. These findings are consistent with previous reports demonstrating the direct association of methylation status to the expressions of E-cad as well as estrogen receptor in cultured cells of esophagus, stomach and breast cancers.18, 20 The reactivation of tumor suppressor genes by demethylating agents, however, was occasionally incomplete in cell lines of breast cancer and prostate,25 suggesting that other mechanisms may also be involved in the expression changes of E-cad.
Cellular migration penetrating surrounding matrix proteins is the first step in the invasive process. In this setting, production and degradation of matrix proteins directly control the behavior of tumor cells under the influence of tumor cell–stroma interactions.36, 37 Adenoid cystic carcinoma cells produce much matrix proteins as well as proteolytic enzymes, which permit the extension of tumor cells into the stroma, diminishing the intercellular adhesive role.38 In vitro, ACC3 cell line also produces basement membrane molecules, including type IV collagen, laminin, heparan sulfate proteoglycan, and fibronectin.6, 28 It has been shown that E-cad methylation density of CpG sites increases when tumor cells are cultured on coated basement membrane molecules.26 Excessive production of extracellular molecules in adenoid cystic carcinoma might thus be associated with a high incidence of E-cad gene methylation. These conditions may as a consequence result in the perturbed expression of E-cad allowing the detachment of tumor cells. This consideration appears to agree with our findings that E-cad methylation was increased with the advancement of the disease and further associated with the reduction of its protein expression in two of our cases (Cases 4 and 26). Further exploration of tumor–stroma interaction will be needed to clarify the precise mechanisms of methylation in the advanced stage, and such approaches will be valuable for the future inhibition of the progression of this intractable disease.
References
Khan AJ, DiGiovanna MP, Ross DA, et al. Adenoid cystic carcinoma: a retrospective clinical review. Int J Cancer (Radiat Oncol Invest) 2001;96:149–158.
Spiro RH, Huvos AG, Strong EW . Adenoid cystic carcinoma: factors influencing survival. Am J Surg 1979;138:579–583.
Seaver PR, Kuehn PG . Adenoid cystic carcinoma of the salivary glands: a study of ninety-three cases. Am J Surg 1979;137:449–455.
Nascimento AG, Amaral ALP, Prado LAF, et al. Adenoid cystic carcinoma of salivary glands: a study of 61 cases with clinicopathologic correlation. Cancer 1986;57:312–319.
Seifert G, Sobin LH . The world health organization's histological classification of salivary gland tumors: a commentary on the second edition. Cancer 1992;70:379–385.
Cheng J, Saku T, Okabe H, et al. Basement membranes in adenoid cystic carcinoma: an immunohistochemical study. Cancer 1992;69:2631–2640.
Shirasuna K, Watatani K, Furusawa H, et al. Biological characterization of pseudocyst-forming cell lines from human adenoid cystic carcinoma of minor salivary gland origin. Cancer Res 1990;50:4139–4145.
Hirohashi S . Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol 1998;153:333–339.
Shiozaki H, Tahara H, Oka H, et al. Expression of immunoreactive E-cadherin adhesion molecules in human cancers. Am J Pathol 1991;139:17–23.
Smith MEF, Pignatelli M . The molecular histology of neoplasia: the role of the cadherin/catenin complex. Histopathology 1997;31:107–111.
Becker K, Atkinson MJ, Reich U, et al. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res 1994;54:3845–3852.
Machado JC, Soares P, Carneiro F, et al. E-cadherin gene mutations provide a genetic basis for the phenotypic divergence of mixed gastric carcinomas. Lab Invest 1999;79:459–465.
Muta H, Noguchi M, Kanai Y, et al. E-cadherin gene mutations in signet ring cell carcinoma of the stomach. Jpn J Cancer Res 1996;87:843–848.
Oda T, Kanai Y, Oyama T, et al. E-cadherin gene mutations in human gastric carcinoma cell lines. Proc Natl Acad Sci USA 1994;91:1858–1862.
Tamura G, Sakata K, Nishizuka S, et al. Inactivation of the E-cadherin gene in primary gastric carcinomas and gastric carcinoma cell lines. Jpn J Cancer Res 1996;87:1153–1159.
Hiraguri S, Godfrey T, Nakamura H, et al. Mechanisms of inactivation of E-cadherin in breast cancer cell lines. Cancer Res 1998;58:1972–1977.
Franchi A, Gallo O, Bocciolini C, et al. Reduced E-cadherin expression correlates with unfavorable prognosis in adenoid cystic carcinoma of salivary glands of the oral cavity. Am J Clin Pathol 1999;111:43–50.
Yoshiura K, Kanai Y, Ochiai A, et al. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA 1995;92:7416–7419.
Gonzalgo ML, Hayashida T, Bender CM, et al. The role of DNA methylation in expression of the p19/p16 locus in human bladder cancer cell lines. Cancer Res 1998;58:1245–1252.
Ferguson AT, Lapidus RG, Baylin SB, et al. Demethylation of the estrogen receptor gene in estrogen receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res 1995;55:2279–2283.
Maruya S, Kurotaki H, Shimoyama N, et al. Expression of p16 protein and hypermethylation status of its promoter gene in adenoid cystic carcinoma of the head and neck. ORL 2003;65:26–32.
Tamura G, Yin J, Wang S, et al. E-cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst 2000;92:569–573.
Kanai Y, Ushijima S, Hui AM, et al. The E-cadherin gene is silenced by CpG methylation in human hepatocellular carcinomas. Int J Cancer 1997;71:355–359.
Graff JR, Greenberg VE, Herman JG, et al. Distinct patterns of E-cadherin CpG island methylation in papillary, follicular, Hurthle's cell, and poorly differentiated human thyroid carcinoma. Cancer Res 1998;58:2063–2066.
Graff JR, Herman JG, Lapidus RG, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res 1995;55:5195–5199.
Graff JR, Gabrielson E, Fujii H, et al. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem 2000;275:2727–2732.
Szanto PA, Luna MA, Tortoledo E, et al. Histologic grading of adenoid cystic carcinoma of the salivary glands. Cancer 1984;54:1062–1069.
Cheng J, Irie T, Munakata R, et al. Biosynthesis of basement membrane molecules by salivary adenoid cystic carcinoma cells: an immunofluorescence and confocal microscopic study. Virchows Arch 1995; 426:577–586.
Goelz SE, Hamilton SR, Vogelstein B . Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun 1985;130:118–126.
Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996;93:9821–9826.
De Medina SG, Popov Z, Chopin DK, et al. Relationship between E-cadherin and fibroblast growth factor receptor 2b expression in bladder carcinomas. Oncogene 1999;18:5722–5726.
Nass SJ, Herman JG, Gabrielson E, et al. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res 2000;60:4346–4348.
Dong SM, Kim HS, Rha SH, et al. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res 2001;7:1982–1986.
Corn PG, Heath EI, Heitmiller R, et al. Frequent hypermethylation of the 5′ CpG island of E-cadherin in esophageal adenocarcinoma. Clin Cancer Res 2001;7:2765–2769.
Bornman DM, Mathew S, Alsruhe J, et al. Methylation of the E-cadherin gene in bladder neoplasia and in normal urothelial epithelium from elderly individuals. Am J Pathol 2001;159:831–835.
Nakagawa H, Yagihashi S . Expression of type IV collagen and its degrading enzymes in squamous cell carcinoma of lung. Jpn J Cancer Res 1994;85:934–938.
Levy AT, Cioce V, Sobel ME, et al. Increased expression of the Mr 72,000 type IV collagenase in human colonic adenocarcinoma. Cancer Res 1991;51:439–444.
Shirasuna K, Saka M, Hayashido Y, et al. Extracellular matrix production and degradation by adenoid cystic carcinoma cells: participation of plasminogen activator and its inhibitor in matrix degradation. Cancer Res 1993;53:147–152.
Acknowledgements
We are greatly indebted to Ms Nozomi Masuta, Mr Masakazu Kobayashi for their expert technical assistance, Dr Norihiko Shimoyama in Hakodate Municipal Hospital, and Dr Mitsuomi Kaimori in Aomori Prefecture Hospital for providing cases of ACC. We also greatly appreciate Professor Bill Orr in the Department of Pathology at the University of Manitoba, Canada for his kind suggestions and comments on this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Maruya, Si., Kurotaki, H., Wada, R. et al. Promoter methylation and protein expression of the E-cadherin gene in the clinicopathologic assessment of adenoid cystic carcinoma. Mod Pathol 17, 637–645 (2004). https://doi.org/10.1038/modpathol.3800104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800104
Keywords
This article is cited by
-
Adenoid Cystic Carcinoma and Polymorphus Adenocarcinoma: An Immunohistochemical Comparison
Iranian Journal of Science and Technology, Transactions A: Science (2021)
-
Expression of integrin-linked kinase in adenoid cystic carcinoma of salivary glands correlates with epithelial–mesenchymal transition markers and tumor progression
Medical Oncology (2013)
-
Differential expression of topoisomerase IIα protein in salivary gland carcinomas: histogenetic and prognostic implications
BMC Cancer (2009)
-
Effects of small interfering RNA targeting thymidylate synthase on survival of ACC3 cells from salivary adenoid cystic carcinoma
BMC Cancer (2008)