Abstract
The recent development of tissue microarray technology has potentiated large-scale retrospective cohort studies using archival formalin-fixed, paraffin-embedded tissues. A major obstacle to broad acceptance of microarrays is that they reduce the amount of tissue analyzed from a whole tissue section to a disk, 0.6 mm in diameter, that may not be representative of the protein expression patterns of the entire tumor. In this study, we examine the number to disks required to adequately represent the expression of three common antigens in invasive breast carcinoma—estrogen receptor, progesterone receptor, and the Her2/neu oncogene—in 38 cases of invasive breast carcinoma. We compared the staining of 2 to 10 microarray disks and the whole tissue sections from which they were derived and determined that analysis of two disks is comparable to analysis of a whole tissue section in more than 95% of cases. To evaluate the potential for using archival tissue in such arrays, we created a breast cancer microarray of 8 to 11 cases from each decade beginning in 1932 to the present day and evaluated the antigenicity of these markers and others. This array demonstrates that many proteins retain their antigenicity for more than 60 years, thus validating their study on archival tissues. We conclude that the tissue microarray technique, with 2-fold redundancy, is a valuable and accurate method for analysis of protein expression in large archival cohorts.
Similar content being viewed by others
Introduction
For over a century, tissue has been preserved in formalin and embedded in paraffin for sectioning before microscopic examination. This method has become the standard method of histopathologic analysis (Wright, 1985). Sampling tissue at the rate of 1 section per cubic cm of tumor became the standard of care for minimal representative sampling of a lesion. Given a tumor 1 cm in cubic diameter, the standard 5-μm-thick tissue section represents 0.05% of the tumor.
In 1998, Kononen and colleagues in the lab of Ollie Kallioniemi invented a mechanism for examining several histologic sections at one time by arraying them in a paraffin block (Kononen et al, 1998). These tissue microarrays are assembled by taking core needle “biopsies” of pre-existing paraffin-embedded tissues and re-embedding them in an arrayed “master” block. In this way, tissue from hundreds of specimens can be represented on a single paraffin block that can be analyzed using a variety of techniques, including immunohistochemistry and in situ fluorescence hybridization (FISH) (Bubendorf et al, 1999; Kononen et al, 1998; Moch et al, 1999; Mucci et al, 2000; Perrone et al, 2000; Schraml et al, 1999). In contrast to traditional techniques, which require the processing and staining of hundreds of slides, microarray technology enables the study of an entire cohort of cases by analyzing just one (or a few) master slide(s). Microarray analysis has the added advantage that all specimens are processed at one time using identical conditions. Furthermore, it markedly reduces the amount of archival tissue required for a particular study, thus preserving ample remaining tissue for other research or diagnostic needs (Mills et al, 1995).
Perhaps the greatest potential disadvantage of this new technology is that it reduces the amount of tumor analyzed. Each disk of tissue represents only about 1.4×106 cubic microns, or about 2 to 3 high-power fields. Quantitatively, this represents about 0.3% of the tissue currently considered “representative.” Although Kallioniemi’s group has shown data from large analyses suggesting there is no difference in outcome when analyzing any one spot of a group of four, to our knowledge this data is unpublished. Thus, one goal of this validation study is to determine how many tissue disks are required to adequately represent the expression of a particular antigen by a tumor.
The second goal of this validation study is to determine antigen survival because this new technique has the potential to examine large cohorts of patients with long-term follow-up. Specifically, we were interested in determining whether archival tissues retain their antigenicity despite decades of storage as paraffin blocks. There is some evidence to suggest that such tissues remain antigenically intact (Ibrahim et al, 1997; Shibata et al, 1988). On the other hand, although paraffin should protect the tissue from oxidation or other damage, there is evidence that once tissues are sectioned they are subject to rapid loss of antigenicity (Jacobs et al, 1996; Shin et al, 1997). This question is also revisited because maximal sectioning of microarrays requires many array sections to be cut at one time, even if they are not analyzed within the same day or week. Both of these questions are addressed as the second goal of this validation study.
To assess these potential limitations of this tissue microarray technique, we have constructed a series of specially designed microarray blocks. They include a high-redundancy array (2- to 10-fold) of 38 invasive breast cancers and a microarray using invasive breast cancers from each decade, beginning with the 1930s. Standard immunohistochemical analyses were performed using several common antigen/antibody pairs including estrogen receptor (ER), progesterone receptor (PR), the Her2/neu oncogene (HER2), Ki-67, and cytokeratin. This study suggests that two needle cores adequately represent the antigen expression on a whole tissue section with greater than 95% accuracy and that most proteins retain their antigenicity for up to 70 years.
Results and Discussion
To determine the number of tissue disks required to obtain a result equivalent to a conventional tissue section, we constructed an array with 10 cores from each of 38 cases of invasive breast cancer. Disks were scored if at least 10% of the disk area contained tumor. Because of technical limitations (described below), most cases had fewer that 10 scorable disks for each antibody tested. However, most cases had more than 6 disks with sufficient tumor for evaluation. A minimum of 2 usable disks was required to be included in the analysis, corresponding to 30 to 35 cases per slide.
Microarrays and the original block from which they were derived were stained for estrogen receptor, progesterone receptor, and Her2/neu. These antigens were chosen, in part because they are routinely performed on diagnostic specimens, and also because their expression from place to place within individual breast cancers is regarded as significantly heterogeneous. (Brennan et al, 1979; Layfield et al, 1998; Rosen et al, 1977; van Netten et al, 1985). In our cohort the percentage of ER-, PR-, and HER2-positive cases, based on whole-section staining, was 68%, 56%, and 24%, respectively. Analysis of estrogen receptors revealed that, in 35 of 36 cases, the average staining of the microarray disks agreed with the whole-section analysis (Table 1). In one case (Case 6), the whole tissue section was graded as borderline-negative (5% staining), whereas the array cores averaged as positive (28% staining, 9 cores analyzed). Of the nine cores, three were graded as negative and four as positive. Eight other cases exhibited nonuniform cores (ie, some negative and some positive); however, in each of these cases, the average staining pattern agreed with the whole section.
Analysis of the progesterone receptor demonstrated similar results. Microarray analysis of 1 of 35 cases disagreed with the whole-section stain (40% average of 9 cores vs 5% whole-section staining; Table 1, Case 11). Five cases contained both positive and negative cores, but the average staining pattern agreed with the whole section in each.
Expression of HER2 showed slightly more variability (Table 1). Of 34 cases, one (Case 14) was discrepant with the whole section (1.38 vs 2, respectively, 8 cores examined). Nine cases contained both positive and negative cores, but again the average staining correlated with that of the whole section (Table 1).
Several studies have shown that certain proteins are preferentially expressed on cells from the leading edge versus those from the center of the tumor mass (Brooks and Leathem, 1995; Cardillo et al, 1997). To address this issue, we took half of our cores from the periphery of the tumor and half from the center. In seven cases, 3 ER, 2 PR, and 2 HER2, there was discrepant staining between these two areas (Table 1 and Fig. 1). This result suggests that cores from both the tumor edge and tumor center may be required for adequate representation of a tumor's immunohistochemical profile.
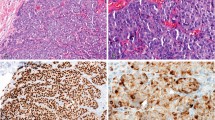
An example of a breast carcinoma expressing high levels of progesterone receptor (PR) at the periphery but not in the center of the lesion. Immunohistochemical analysis of PR is shown in panel A (diaminobenzidine [DAB] chromagen). An hematoxylin and eosin (H&E) section taken from the block after tissue cores were removed is shown in panel B. Note that cores (holes) taken from the tumor periphery (arrowheads) are in areas of intense staining, whereas those in the tumor center (circles) are not.
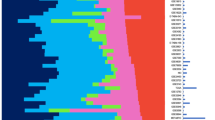
We then determined how many cores it takes to adequately represent a tumor. To do this we analyzed each disk separately and graded it as either positive or negative. The average chance of correctly calling each case was calculated. In the majority of cases, staining was uniform across all the disks and consequently the chance of adequately representing the tumor in one punch was 100%. In the cases that contained mixed cores (Table 1), the chances of being correct ranged from 90% to 42.8% per disk. Despite the fact that the average score of three cases (Cases 6, 11, and 14; Table 1) failed to match that of the whole sections from which they came, all three contained mixed positive and negative disks. Consequently, the theoretical maximum of correctly calling all cases (eg, with an infinite number of cores) remained 100%. The results of this analysis demonstrate that, despite the variability of antigen expression between cores, analysis of a single readable disk would match the staining pattern of an entire section more than 90% of the time (Fig. 2). Analysis of two disks achieve greater than 95% representation, thus attaining the commonly accepted level of statistical significance.
The chance of 1 to 10 microarray disks correctly representing the staining of a whole tissue section. Disks from breast carcinoma cases were stained for estrogen receptor (ER), PR, and Her2/neu oncogene (HER2). The staining pattern of each disk was independently analyzed and the chance of one disk matching the staining pattern of the whole tissue section from which it came was determined. The chance of 2 to 10 disks being representative was directly extrapolated and plotted.
The question of how many core punches it takes to reliably achieve two readable disks in each case is affected by several factors. Foremost among these is the technical expertise of the individual constructing the array blocks and slides. In our laboratory, one person performed these tasks, and the number of usable disks climbed with each successive array constructed. Presently our rate of usable disks exceeds 95% (data not shown). Other factors include the ability to identify areas of tumor distinct from desmoplastic stroma, normal epithelium, and/or in situ carcinoma. Although some histologic detail is retained in microarray disks, it can be difficult to distinguish certain types of in situ from invasive carcinoma. Furthermore, in situ carcinoma often has an antigen profile different from coincident invasive carcinoma. In contrast to certain specimens (eg, colon carcinoma and melanoma), in situ breast carcinoma is often admixed with the invasive component, increasing the chances of spurious readings. To reduce this confounding effect, we carefully outlined areas of invasive carcinoma distinct from the in situ component. This process was performed by a technician and verified by a pathologist. Finally, the thickness (depth) of the original embedded tissue sections can affect the number of usable slides able to be cut from the master array block. Therefore, efforts were made to select blocks that had not been previously cut numerous times (ie, for other studies or diagnostic immunostains).
Given this data and our experience, we have developed a standard procedure in which each tumor block is punched three times in various regions of the tumor mass, including both the leading edge and the tumor center. This process ensures that at least two, and in most cases three, punches are available for evaluation, resulting in adequate representation of the whole-section staining pattern in more than 95% of cases. Although expression of estrogen receptor, progesterone receptor, and HER2/neu in breast cancer is regarded as particularly heterogeneous, it is possible that other antigens in other tumor types may exhibit even greater heterogeneity. Consequently, similar analyses may be required to validate the use of tissue microarrays in other tumor types.
The second major issue that this study addresses is antigen durability. Because of the potential to use archival tissues to generate large cohorts of patients with long-term follow-up, we were interested in determining how long paraffin-embedded tissue blocks retained their antigenicity, both before and after sectioning. The ability to use archival tissues from several decades is appealing for several reasons. First, in rare tumors, such a time frame may be required to achieve sufficiently large numbers of tumors to analyze. Second, combinations of cases from various decades may help negate treatment differences that can affect the follow-up data of cohorts from shorter time periods. Third, longer-term patient follow-up ensures that late recurrences are not missed. This is particularly important in breast cancer where tumors can recur decades after their initial presentation.
The preservation of antigens in fixed tissue specimens is poorly understood and somewhat dependent upon differences in fixation, paraffin embedding, block storage, and antigen retrieval. In previous studies, we have routinely used sections that were up to 15 years old with no perceptible change in antigenicity (Camp et al, 1999; Dillon et al, 1998; Ghoussoub et al, 1998) using a standard “antigen retrieval” protocol (Norton et al, 1994). This study presents a quantitative analysis of this issue.
The Yale University Department of Pathology has retained paraffin-embedded tissue since 1932. This archive includes over 14,000 cases of breast cancer, approximately 40% of which are from before 1983. To assess the potential use of this archive, we developed a microarray of breast cancer cases from each decade beginning in 1932. The microarray was constructed using three cores from each case, and only those cases in which there were at least two readable disks were analyzed. The array was analyzed for the expression of breast-specific antigens including ER, PR, HER2. The array was also examined for keratin expression, Ki-67 expression, and DAPI staining. The number of positively staining cases for the breast-specific antigens from each decade is shown in Figure 3. Because immunohistochemical analysis of these tumors was not performed at the time of diagnosis (most were diagnosed before the development of such techniques), nonstaining cases may either be true negatives or cases in which the antigenicity of the specimen is not preserved. However, the presence of positive cases within all decades represented on the microarray suggests that many proteins are antigenically retrievable using standard immunohistochemical techniques on tissues stored for more than 60 years.
The percentage of cases expressing ER, PR, and HER2 per decade. Microarrays of 8 to 11 cases from each decade beginning in 1932 were analyzed for their expression of ER, PR, and HER2, and the percentage of positive cases per decade was plotted. For each antigen, the number of positive cases from each year was compared to the remaining years using a Chi-square test, with the results expressed as a p value at the end of each bar.
Comparisons across decades reveal that there are no dramatic shifts in the levels of antigen expression, with the exception of ER, which was identified on only one case in the 1932 cohort (Fig. 3). The lack of ER staining in 1932 cases achieved statistical significance when compared with cases from the remaining years (p = 0.0002). This result may represent a true phenomenon, ie, that tumors in the 1930s were discovered later, were more aggressive, and consequently expressed less ER. Alternatively, it may represent a lack of ER antigenicity by most of the tissue from that era. Given the relatively small sample number from each year represented in the array, it was not possible to distinguish between these two possibilities. However, the goal of this array was to determine whether, in a given year, a significant percentage of cases stained positively for a particular antigen, thereby demonstrating that tissue from that year was antigenically viable. The statistically significant decreased staining of ER in cases from 1932 would call into question the use of that antibody on cases from that era. The signal intensity of ER, PR, and HER2/neu, as well as other antigens (cytokeratin and Ki-67), did not appear diminished on the early versus late cases (data not shown). Interestingly, nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI) was diminished in cases before 1952.
Our results suggest that in most cases, archival tissue from up to 68 years ago is suitable for immunohistochemical analysis. In some cases, the absence of positive staining prior to a given time (eg, ER) may warrant the exclusion of earlier cases. Because fixation protocols, tissue processing, and block storage conditions vary over time, we suggest that decade-style arrays play an important role in validating the use of particular antibodies on archival tissues. Furthermore, because tissue preparations will also vary from institution to institution, we suggest that each institution prepare such arrays to validate the use of their own archival material.
In summary, this tissue microarray technique appears to be fairly robust with respect to sampling. One or two disks per case result in outcomes that are 95% similar to those achieved using conventional tissue sections. Furthermore, this study suggests that good results may be achieved using archival specimens to obtain long term follow-up.
Materials and Methods
Formalin-fixed, paraffin-embedded tissue blocks containing invasive breast cancers were retrieved, along with their corresponding hematoxylin and eosin (H&E)-stained slides, from the archives of the Yale University Department of Pathology (New Haven, Connecticut). The fixation, tissue processing, and early storage protocols of these tissues are unknown; however, they are likely to be highly variable. In general, blocks were stored under ambient conditions in an approximate temperature range of 18 to 37° C. Paraffin blocks created before the use of plastic cassettes (circa 1960) were melted and re-embedded, and new H&E sections were cut.
Cases of invasive breast carcinoma from each decade, from 1932 to 1999, were chosen. Cases were selected sequentially beginning with the first case from each year for which tissue was available. Areas of invasive carcinoma, distinct from in situ components and normal epithelium, were identified on the stained slides by a technician (LAC) and a pathologist (RLC) and marked for subsequent analysis. The paraffin-embedded tissue corresponding to these areas was cored 10 times: 5 times from the periphery and 5 from the center of the lesion. In 29 of 38 cases, this represented the actual tumor edge and center; however, in the remaining 9 cases, the assayed tissue block contained tumor without surrounding normal tissue and only the periphery and center of the block could be represented. Cores were transferred to a recipient paraffin block using the technique described by Kononen et al (1998) using a Tissue Microarrayer (Beecher Instruments, Silver Spring, Maryland).
Microarray slides were processed within 2 weeks of cutting, except as indicated. Both microarray and whole-section slides were stained using techniques and reagents from DAKO (Carpinteria, California). In brief, slides were deparaffinized and antigens were retrieved using microwave antigen retrieval (Katoh et al, 1997). Slides were then stained with primary antibodies including monoclonal antibodies to ER and PR as well as polyclonal rabbit antibody to HER2. Slides were then incubated with mouse- or rabbit-specific secondary antibodies covalently linked to dextran polymers containing multiple peroxidase molecules (DAKO EnVision® System and HercepTest®, DAKO) and developed with diaminobenzidine (DAB). All slides were processed using a DAKO automatic stainer. Other antibodies used in this study include Ki-67 (BD Transduction Laboratories, Lexington, Kentucky) and polyclonal rabbit anticytokeratin (Zymed, South San Francisco, California). To prevent oxidation of array slides after sectioning, the slides were deparaffinized, dried, and dipped in a layer of paraffin. Immediately before staining, the paraffin was removed by incubating in a 60° C incubator for 30 minutes and following with several washes in xylene.
Levels of ER, PR, and HER2 staining were analyzed according to the manufacturers instructions in a manner identical to that used for routine diagnostic specimens (at Yale New Haven Hospital, New Haven, Connecticut). In brief, ER and PR levels were assessed by the number of positively stained nuclei, with a score greater than 10% indicating a positive result. HER2 staining was assessed on a 0- to 3-point scale, with scores greater than 1.5 designated as positive. Disks from the same case were independently assessed and given a separate score. Disk scores from the same tumor were either averaged together to produce a single score or expressed as a ratio of positive to negative cases, depending on the analysis.
References
Brennan MJ, Donegan WL, and Appleby DE (1979). The variability of estrogen receptors in metastatic breast cancer. Am J Surg 137: 260–262.
Brooks SA and Leathem AJ (1995). Expression of the CD15 antigen (Lewis x) in breast cancer. Histochem J 27: 689–693.
Bubendorf L, Kolmer M, Kononen J, Koivisto P, Mousses S, Chen Y, Mahlamaki E, Schraml P, Moch H, Willi N, Elkahloun AG, Pretlow TG, Gasser TC, Mihatsch MJ, Sauter G, and Kallioniemi OP (1999). Hormone therapy failure in human prostate cancer: Analysis by complementary DNA and tissue microarrays. J Natl Cancer Inst 91: 1758–1764.
Camp RL, Rimm EB, and Rimm DL (1999). Met expression is associated with poor outcome in patients with axillary lymph node negative breast carcinoma. Cancer 86: 2259–2265.
Cardillo MR, Yap E, and Castagna G (1997). Molecular genetic analysis of TGF beta1 in breast cancer. J Exp Clin Cancer Res 16: 57–63.
Dillon DA, Daquila T, Reynolds AB, Fearon ER, and Rimm DL (1998). The expression of P120ctn protein in breast cancer is independent of alpha- and beta-catenin and E-cadherin. Am J Pathol 152: 75–82.
Ghoussoub RAD, Dillon DA, Daquila T, Rimm EB, Fearon ER, and Rimm DL (1998). Expression of c-met is a strong independent prognostic factor in breast carcinoma. Cancer 82: 1513–1520.
Ibrahim SO, Johannessen AC, Vasstrand EN, Lillehaug JR, and Nilsen R (1997). Immunohistochemical detection of p53 in archival formalin-fixed tissues of lip and intraoral squamous cell carcinomas from Norway. APMIS 105: 757–764.
Jacobs TW, Prioleau JE, Stillman IE, and Schnitt SJ (1996). Loss of tumor marker-immunostaining intensity on stored paraffin slides of breast cancer [see comments]. J Natl Cancer Inst 88: 1054–1059.
Katoh AK, Stemmler N, Specht S, and D'Amico F (1997). Immunoperoxidase staining for estrogen and progesterone receptors in archival formalin fixed, paraffin embedded breast carcinomas after microwave antigen retrieval. Biotech Histochem 72: 291–298.
Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, and Kallioniemi OP (1998). Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4: 844–847.
Layfield LJ, Saria E, Mooney EE, Liu K, and Dodge RR (1998). Tissue heterogeneity of immunohistochemically detected estrogen receptor. Implications for image analysis quantification. Am J Clin Pathol 110: 758–764.
Mills SE, Fechner RE, Frierson HF, Kempson RL, Wick MR, Dehner LP, Swanson PE, and Humphrey PA (1995). Guardians of the wax and the patient. Am J Clin Pathol 104: 365–367.
Moch H, Schraml P, Bubendorf L, Mirlacher M, Kononen J, Gasser T, Mihatsch MJ, Kallioniemi OP, and Sauter G (1999). High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol 154: 981–986.
Mucci NR, Akdas G, Manely S, and Rubin MA (2000). Neuroendocrine expression in metastatic prostate cancer: Evaluation of high throughput tissue microarrays to detect heterogeneous protein expression. Hum Pathol 31: 406–414.
Norton A, Jordan S, and Yeomans P (1994). Brief high-temperature heat denaturation (pressure cooking): A simple and effective method of antigen retrieval for routinely processed specimens. J Pathol 173: 371–379.
Perrone EE, Theoharis C, Mucci NR, Hayasaka S, Taylor JM, Cooney KA, and Rubin MA (2000). Tissue microarray assessment of prostate cancer tumor proliferation in African-American and white men. J Natl Cancer Inst 92: 937–939.
Rosen PP, Menendez-Botet CJ, Urban JA, Fracchia A, and Schwartz MK (1977). Estrogen receptor protein (ERP) in multiple tumor specimens from individual patients with breast cancer. Cancer 39: 2194–2200.
Schraml P, Kononen J, Bubendorf L, Moch H, Bissig H, Nocito A, Mihatsch MJ, Kallioniemi OP, and Sauter G (1999). Tissue microarrays for gene amplification surveys in many different tumor types. Clin Cancer Res 5: 1966–1975.
Shibata D, Martin WJ, and Arnheim N (1988). Analysis of DNA sequences in forty-year-old paraffin-embedded thin-tissue sections: A bridge between molecular biology and classical histology. Cancer Res 48: 4564–4566.
Shin HJ, Kalapurakal SK, Lee JJ, Ro JY, Hong WK, and Lee JS (1997). Comparison of p53 immunoreactivity in fresh-cut versus stored slides with and without microwave heating. Mod Pathol 10: 224–230.
van Netten JP, Algard FT, Coy P, Carlyle SJ, Brigden ML, Thornton KR, Peter S, Fraser T, and To MP (1985). Heterogeneous estrogen receptor levels detected via multiple microsamples from individual breast cancers. Cancer 56: 2019–2024.
Wright JR Jr (1985). The development of the frozen section technique, the evolution of surgical biopsy, and the origins of surgical pathology. Bull Hist Med 59: 295–326.
Acknowledgements
The authors wish to thank Mary Helie for her help with tissue immunohistochemistry and John Sinard and Art Belanger for their help in assembling a comprehensive database of pathology specimens from 1915 to 1983.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is supported by grants from the William and Catherine Weldon Donaghue Foundation for Medical Research and the Argall L. and Anna G. Hull Fund, NIH R21-CA81333.
Rights and permissions
About this article
Cite this article
Camp, R., Charette, L. & Rimm, D. Validation of Tissue Microarray Technology in Breast Carcinoma. Lab Invest 80, 1943–1949 (2000). https://doi.org/10.1038/labinvest.3780204
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3780204
This article is cited by
-
“Evaluation of ROS1 expression and rearrangements in a large cohort of early-stage lung cancer”
Diagnostic Pathology (2023)
-
Co-expression of cancer stem cell markers, SALL4/ALDH1A1, is associated with tumor aggressiveness and poor survival in patients with serous ovarian carcinoma
Journal of Ovarian Research (2022)
-
Development of an immunohistochemical assay for Siglec-15
Laboratory Investigation (2022)
-
Models that combine transcriptomic with spatial protein information exceed the predictive value for either single modality
npj Precision Oncology (2021)
-
DTX3 copy number increase in breast cancer: a study of associations to molecular subtype, proliferation and prognosis
Breast Cancer Research and Treatment (2021)