Abstract
The comparison of gene expression measurements obtained with different technical approaches is of substantial interest in order to clarify whether interplatform differences may conceal biologically significant information. To address this concern, we analyzed gene expression in a set of head and neck squamous cell carcinoma patients, using both spotted oligonucleotide microarrays made from a large collection of 70-mer probes and commercial arrays produced by in situ synthesis of sets of multiple 25-mer oligonucleotides per gene. Expression measurements were compared for 4425 genes represented on both platforms, which revealed strong correlations between the corresponding data sets. Of note, a global tendency towards smaller absolute ratios was observed when using the 70-mer probes. Real-time quantitative reverse transcription PCR measurements were conducted to verify expression ratios for a subset of genes and achieved good agreement regarding both array platforms. In conclusion, similar profiles of relative gene expression were obtained using arrays of either single 70-mer or multiple short 25-mer oligonucleotide probes per gene. Although qualitative assessments of the expression of individual genes have to be made with caution, our results indicate that the comparison of gene expression profiles generated on these platforms will help to discover disease-related gene signatures in general.
Similar content being viewed by others
Main
Today, researchers can choose from a broad variety of methods for global transcriptional profiling. Among the different technical approaches, microarray technology has gained a premier position. In principle, microarrays can be produced either by robotic printing (‘spotting’) of DNA on a chemically modified glass surface,1 or by in situ synthesis of oligonucleotides via custom phosporamidite chemistry using either photolithography on a silane-reacted quartz substrate2 or ink-jet technology on a hydrophobic glass support.3
Spotted arrays usually contain cDNA-specific PCR amplicons (cDNA arrays), ranging from several hundred to a few thousand basepairs in size. Generally, no more than one amplicon is used to probe a given gene. Although they are technically challenging and require both optimized protocols4 and workflow,5 cDNA arrays are typically produced by individual research groups or core facilities. Alternatively, they can be purchased from several commercial suppliers. But after the discovery of frequent discrepancies in the annotation of cDNA clones,6 investigators began to realize potential drawbacks of this highly advocated technology. In situ synthesis of oligonucleotide probes requires sophisticated equipment for photolithography and solid phase chemistry, which is usually too complex and elaborate for an academic environment. A widespread commercial implementation of this technology is the Affymetrix GeneChip platform,2 which currently uses 11–16 pairs (11 for the arrays used in this study) of perfect-match and single-base-mismatch 25-mer oligonucleotides for each gene. Recently, large collections of longer oligonucleotides (50–80 bases), produced by established suppliers using conventional phosphoramidite chemistry, have become increasingly popular as probes for spotted DNA arrays. Technical advantages of oligonucleotide arrays include a constant DNA concentration across all spots and biophysically optimized sequences, reducing secondary structures, avoiding repetitive sequence motives and providing a fixed range for both Tm and length. All this accounts for more uniform, stable and predictable hybridization conditions. The overall costs for long oligonucleotide arrays will often be lower when labor and other costs associated with cDNA libraries, such as replication, amplification or sequence verification, are regarded.
Considering this diversity of approaches and the resulting technical differences, researchers are highly interested in the general accuracy and reliability of microarray data and the cross-platform comparability. Several independent methods like Northern blotting or real-time quantitative reverse transcription-PCR (RQ-PCR) have been used to validate microarray results for a small number of transcripts. Generally, there was a good agreement between the corresponding values, affirming the ability to accurately profile gene expression with array-based approaches.
Former studies also compared global expression measurements between cDNA arrays and short oligonucleotide arrays7, 8 or SSH.9 Recently, Barczak et al10 compared results between spotted arrays of 70–mer oligonucleotides and in situ synthesized Affymetrix GeneChip arrays. Using RNA of a cell line and a commercial reference RNA, they found strong correlations of the corresponding data sets. Despite these studies clarifying some fundamental questions, there still remains considerable uncertainty regarding the comparability of data from clinical specimens. As this lack of understanding constitutes a barrier, which keeps researchers from an immense amount of potentially valuable information (via efficient integration of microarray data generated on different array platforms), we conducted a comparison with tumor samples from clinical practice, which evaluates cross-platform reproducibility in a practical setting.
Materials and methods
Experimental Design
To assess the degree of concordance between expression profiles obtained with either spotted oligonucleotide microarrays made from a large collection of 70-mer probes or commercial arrays produced by in situ synthesis of sets of multiple 25-mer oligonucleotides per gene, we analyzed relative gene expression in a set of six human head and neck squamous cell carcinoma (HNSCC) samples vs either healthy control mucosa (n=4) or lymph node metastases (n=2) of the respective patients as the reference (Table 1). For the spotted 70-mer arrays, relative expression levels were calculated by averaging the normalized log2-ratios of two replicate two-color hybridizations per patient, one performed with inverse assignment of fluorophores (dye swap). This procedure was used to eliminate dye-related signal correlation bias.11, 12 For the commercial 25-mer arrays, relative expression levels were derived by subtracting normalized log2-transformed probe-level data (fluorescence intensities) of two single-color hybridizations per patient, corresponding to the respective tumor and reference tissue.
Patient Characteristics and Tissue Samples
Tissue samples from six patients were obtained during 1998–2002 from patients undergoing surgical resection at the Department of Otorhinolaryngology, JW-Goethe Universität Frankfurt. All cases were diagnosed histopathologically as HNSCC and staged according to the TNM classification of malignant tumors,13 based on criteria recommended by the ‘Union International contre le Cancer’ (UICC) (Table 1). The study protocol was approved by the local ethics committee after obtaining the patients' informed consent to participate in the study, and was processed anonymously. Grade 2 HNSCC specimens, corresponding healthy control mucosa surrounding the tumor and lymph node metastases were surgically resected, immediately frozen in liquid nitrogen and stored at −80°C. The neoplastic specimens contained >80% tumor tissue and <10% necrotic debris.
RNA Extraction
Frozen tissue samples (30–50 mg) were combined with 1 ml Trizol (Invitrogen, Karlsruhe, Germany) and dispersed using an Ultra-Turrax T25 tissue homogenizer (IKA Werke, Staufen, Germany). Total RNA was extracted according to the recommendations given by the Trizol protocol and further purified on RNeasy Mini spin columns (Qiagen, Hilden, Germany). Integrity and purity of total RNA were assessed on a Bioanalyzer 2100 (Agilent Technologies, Boeblingen, Germany) using a RNA 6000 Nano LabChip Kit (Agilent) according to the manufacturer's instructions.
Preparation and Postprocessing of Spotted Oligonucleotide Arrays
Synthetic 70-mer oligonucleotides (‘Human Genome Oligo Set Version 2.1’; consisting of 21 329 oligonucleotides representing human genes and transcripts plus 24 controls, as well as ‘Human Genome Oligo Set Version 2.1 Upgrade’, consisting of 5462 human 70-mer probes) were purchased from Operon Technologies (Cologne, Germany) and dissolved in FBNC spotting buffer (Formamide, Betaine, NitroCellulose)4 at 40 μM, using a MiniTrak robotic liquid handling system (Perkin Elmer, Rodgau-Juegesheim, Germany). DNA spotting was performed in duplicates on Nexterion Slide E epoxysilane coated slides (SCHOTT Nexterion, Mainz, Germany) using a VersArray ChipWriter Pro microarrayer (Bio-Rad, Munich, Germany) equipped with Stealth SMP3 Micro Spotting Pins (Telechem, Sunnyvale, USA). Spot centers were 129 μm apart. DNA adhesion to the glass surface was accomplished by 1 h incubation at 60°C, followed by UV irradiation (2 × 120 mJ/cm2 at 254 nm) in a Stratalinker Model 2400 UV illuminator (Stratagene). Just prior to hybridization, slides were washed for 2 min in 0.2% SDS (w/v), 2 min in ddH2O at room temperature and 2 min in boiling ddH2O (95°C), followed by 3 min centrifugation at 2000 r.p.m.
Target Preparation for Spotted Oligonucleotide Arrays
Fluorescent antisense cDNA was prepared from 2 μg total RNA as described elsewhere.12 Briefly, the mRNA content was linearly amplified by in vitro transcription (‘T7 amplification’). The resulting aRNA was subsequently converted to cDNA and labelled by dye-dUTP incorporation using Klenow fragment.
Hybridization of Spotted Oligonucleotide Arrays
Corresponding dye-labelled cDNA samples were combined and purified on Microcon YM-30 filter columns (Millipore, Eschborn, Germany), as previously described.14 For blocking of repetitive sequence elements, 25 μg C0t-1 DNA (Roche Diagnostics), 25 μg poly-A RNA (Sigma) and 75 μg yeast tRNA (Sigma) were added before the final washing step. Purified, dye-labeled cDNA was mixed with 120 μl UltraHyb hybridization buffer (Ambion), agitated for 30–60 min at 60°C, then for 10 min at 70°C on a thermo mixer and subsequently applied to preheated (60°C) microarrays mounted in a GeneTAC Hybridization Station (Genomic Solutions, Ann Arbor, USA). Hybridizations were performed for 16 h at 42°C with gentle agitation. Thereafter, the arrays were automatically washed at 36°C with (i) 0.5 × SSC, 0.1% (w/v) SDS for 5 min; (ii) 0.05 × SSC, 0.1% (w/v) SDS for 3 min; (iii) 0.05 × SSC for 2 min. Flow time was set to 40 s, respectively. Immediately after completion of the final washing step, the arrays were unmounted, immersed in 0.05 × SSC, 0.1% (w/v) Tween 20 and dried by centrifugation in 50 ml Falcon tubes (30 s at 500, 1000 and 1500 r.p.m., respectively, followed by a final step of 90 s at 2000 r.p.m.).
Scanning, Image Analysis and Data Processing of Spotted Oligonucleotide Arrays
Hybridized microarrays were scanned at 5 μm resolution and variable PMT voltage to obtain maximal signal intensities with <0.1% probe saturation, a count ratio of 0.8–1.2 (Cy5/Cy3) and maximal congruence of histogram curves, using a GenePix 4000B microarray scanner (Axon Instruments, Union City, USA). Subsequent image analysis was performed with the corresponding software GenePix Pro 5.0. Spots not recognized by the software were excluded from further considerations. Result files containing all relevant scan data were further processed using the open source statistical software environment R (http://www.r-project.org)15 together with libraries (packages) of the Bioconductor project (http://www.bioconductor.org).16 Raw fluorescence intensities were normalized applying variance stabilization.17 To eliminate low-quality data, the data points were ranked according to spot homogeneity, as assayed by the ratio of median-to-mean fluorescence intensity, the ratio of spot-to-local background intensity and the standard deviation of the logarithmic ratios (log2 Cy5/Cy3) between spot replicates. Those data points ranked among the lower 20% were removed from the data set. Genes that could not be quantified in more than 33% of all experiments after filtering were excluded as well. To combine the data of dye swap experiments, the log2-transformed intensity ratios of one array were inverted and averaged with the corresponding values of the other array.
Target Preparation, Hybridization and Data Analysis for Affymetrix GeneChip Arrays
Total RNA (5 μg) was used to prepare biotinylated cRNAs for hybridization, following the guidelines given in the Affymetrix GeneChip Expression Analysis Technical Manual.18 cRNA clean-up was performed on RNeasy Mini filters (Qiagen). In all, 10 μg of fragmented, labelled cRNA were hybridized to Affymetrix HG U133A arrays (Affymetrix, Santa Clara, CA, USA) using standard conditions (16 h, 45°C). Arrays were washed and stained in a Fluidics Station 400 (Affymetrix) and scanned on a Gene Array Scanner 2500 (Agilent), as recommended by Affymetrix. Raw fluorescence intensities from all hybridizations were normalized applying variance stabilization17 with additional scaling. Additionally, MAS519 as well as gcRMA20 expression values were calculated.
Matching of Oligonucleotide Probe Sequences
The Bioconductor R package AnnBuilder21 and GenBank accession numbers, provided by Affymetrix and Operon, were used to map probe sequences to corresponding UniGene clusters (build #175). Microarray data were only used if the Affymetrix probe set and the Operon probe corresponded to the same UniGene cluster from the intersection of both platforms (n=4425). For simplicity, if probe sets (Affymetrix) mapped to multiple UniGene clusters or if several probes (Operon) or probe sets (Affymetrix) mapped to the same UniGene, they were excluded from further analyses.
Statistical Analyses
To investigate the linear relationship between data points in Figure 2, regression lines were determined by minimizing the sum of squares of the Euclidean distance of points to the fitted line (‘orthogonal regression’), as there is no clear assignment of dependent and independent variables. Correlations were estimated using the Pearson correlation coefficient together with its 95% confidence interval. An optional filtering procedure additionally excluded those data points considered unreliable as they correspond to probes associated with signal intensities less than two standard deviations above local background for at least one channel of the pair of Operon chips or to probe sets with mean log2 expression values below the median for all probe sets of the pair of GeneChips.10, 22 We chose this strategy in order to extract high-quality data from both array platforms as a sound foundation for quantitative comparisons. More sophisticated filtering based on variance rather than absolute expression levels was not applied due to the deliberate shortage of replicates. Expression ratios of genes with a signal close to the background (low abundance) in only one of the two investigated conditions are clearly significant in a biological context. They were, however, considered less appropriate to this comparative study, as their results were expected to carry an increased and mathematically inevitable degree of variation not caused by characteristics of the investigated platforms.
Scatter plots comparing normalized, log2-transformed expression ratios of spotted long oligonucleotide arrays to ratios obtained with Affymetrix GeneChip short oligonucleotide arrays. For the spotted arrays, normalized ratio data from dye swap experiments were combined. For Affymetrix arrays, the ratios of normalized intensity values from corresponding arrays were used. Hybridized targets were derived from (a) patient 160, (b) patient 171, (c) patient 173, (d) patient 180, (e) patient 186 and (f) patient 205. Orthogonal regression analysis was performed to derive the regression lines shown in black (unfiltered data) and red (filtered data) as well as their respective linear equations (shown in the lower part of the plots for unfiltered data and in the upper part for filtered data). Pearson's correlation coefficients and their associated 95% confidence intervals are listed as well. Dashed lines through origin with slope 1 are displayed to accentuate the reduced slope. For panels (a)–(f), calculations were based on 3472, 3595, 3600, 3569, 3522 and 3474 data points for the unfiltered data sets as well as 1796, 2011, 1889, 1954, 1816 and 1866 data points for the filtered data sets.
Identification of differentially expressed genes was performed by empirical Bayes inference for paired data.23 Moderated t-statistics, based on shrinkage of the estimated sample variance towards a pooled estimate and corresponding P-values, were calculated using the Bioconductor R package limma.24 P-values were adjusted according to the method proposed by Benjamini and Hochberg25 to control the false discovery rate at a level of 10%. The magnitude of the effects as well as the corresponding P-values are illustrated as volcano plots.26
To remove systematic variation resulting from the different technical approaches of the investigated array platforms or differences in sample handling procedures between the two labs participating in this study, ‘Distance Weighted Discrimination (DWD)’27 was performed on normalized log2-ratios from both array platforms, using Matlab software freely available at https://genome.unc.edu/pubsup/dwd/. Further details about cross-platform adjustment of microarray data can be obtained at 〈http://genome.med.unc.edu:8080/caBIG/DWDNCI60.htm〉 and〈http://genome.med.unc.edu:8080/caBIG/paper1.pdf〉. Identification of differentially expressed genes and DWD were only performed for spots with quantified log2-ratios in all four primary HNSCC vs normal mucosa experiments.
Gene ontology (GO)28 data mining was performed using the GOCharts functionality of the ‘Database for Annotation, Visualization and Integrated Discovery (DAVID)’,29 which is available at http://david.niaid.nih.gov/david/. Overrepresentation analysis was carried out with the software application ‘Expression Analysis Systematic Explorer (EASE)’,30 downloaded from http://david.niaid.nih.gov/david/ease.htm.
Real-Time PCR Analysis
For selected genes (Table 2), changes in mRNA levels detected in microarray experiments were evaluated by reverse transcription (RT) and quantitative real-time PCR analysis, using the iCycler (BioRad, Munich, Germany). In all, 1 μg of total RNA was converted to cDNA using Superscript II reverse transcriptase (Invitrogen) and oligo(dT) primer, according to the manufacturer's specifications. PCR reaction mixtures consisted of 12.5 μl of 2 × iQ™ SYBR® Green Supermix (Abgene, Hamburg, Germany), 0.5 μl of each 10 μM target primer and 1 μl diluted cDNA template (1:10) in a reaction volume of 25 μl. Thermal cycling conditions comprised an initial denaturation step of 15 min at 95°C, 40 cycles of 30 s at 95°C and 30 s variable annealing/elongation temperature, depending on the respective set of target primers. dsDNA-specific fluorescence was measured at the end of each extension phase. Product-specific amplification was confirmed by a melting curve analysis. The relative expression ratio (R) of a target gene was calculated using the equation

based on its real-time PCR efficiencies (E) and the crossing point (CP) differences of sample vs a control, and expressed in comparison to a reference gene.31 The target gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Accession Numbers
All relevant data from this study are available from GEO32 (http://www.ncbi.nlm.nih.gov/geo) under the accession numbers GPL96 and GPL1384 (for the array platform), GSM29702-GSM29705, GSM29747-GSM29758, GSM29808-GSM29813, GSM29818 and GSM29820 (for expression data from individual arrays) as well as GSE1722 (for the experimental series).
Results
Probe Matching
We analyzed the gene expression profiles of 12 specimens obtained from six head and neck cancer patients (Table 1). Four primary HNSCC were assayed vs corresponding healthy mucosa and another two primary HNSCC vs corresponding lymph node metastases of the respective patients.
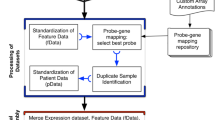
This analysis was performed both on in situ-synthesized Affymetrix HG-U133A arrays, containing 22 283 sets of 25-mer probes, and on spotted long oligonucleotide arrays containing 26 791 70-mer probes of the Operon Human Genome Oligo Set Version 2.1 and Version 2.1 Upgrade. A total of 9867 UniGene clusters were found for the probe sets of the HG-U133A arrays, while 13 604 were retrieved for the Operon arrays, using GenBank accession numbers provided by the manufacturers. A total of 4425 genes were represented on both platforms, as identified by consistent assignment of UniGene clusters to the corresponding probes or probe sets. We used this large set of genes as a basis for comparing expression data from the two array systems (Figure 1).
Intersection of probes or probe sets from the different array platforms. Probe sequences were mapped to UniGene clusters (build #175), based on GenBank accession numbers provided by the manufacturers. A total of 9867 UniGene clusters were found for the probe sets of the HG-U133A arrays, while 13 604 were retrieved for the Operon arrays. A total of 4425 genes were represented on both array types.
Intraplatform Reproducibility of Expression Ratios
For the platform of spotted long oligonucleotide arrays, correlations of expression ratios measured on individual arrays were r=0.99 for identically repeated hybridizations and r=−0.98 for dye swap hybridizations repeated with inverse assignment of fluorophores (data not shown). Similar correlations had been reported for the Affymetrix system.33 Hence, both array platforms provide highly reproducible measurements of gene expression profiles, which is an essential prerequisite for the success of a cross-platform comparison.
Cross-Platform Reproducibility of Expression Ratios
Normalized log2-transformed absolute signal intensities were calculated for the arrays from both platforms using variance stabilization by vsn.17 For GeneChip arrays, log2 expression ratios were obtained by subtracting log2-transformed absolute signal intensities of the two respective arrays from each patient. For the spotted long oligonucleotide array, log2-ratios from two-color dye swap hybridizations were inverted and averaged. To ensure that the observed effects were not due to characteristics of the data processing algorithm, we repeated the analyses of GeneChip arrays using background correction and normalization by gcRMA20 as well as the MAS5 algorithm.19 For all patients, there was a clear correlation between differential expression measurements made with either array type (r=0.56–0.76), and the correlation improved substantially (r=0.61–0.85) when measurements from probes with low-intensity signals were excluded (Figure 2). Except for patients 160 and 186, the respective regression lines all showed a slope clearly smaller than 1, indicating that, on average, absolute log ratios obtained on the Operon long oligonucleotide platform were lower than the corresponding values measured with Affymetrix arrays. The changes in correlation were marginal when gcRMA was used to normalize the GeneChip results. MAS5 yielded lower correlations with unfiltered data, but the results were similar to those of vsn or gcRMA when filtered data were used (Table 3).
Systematic Bias Correction by ‘DWD’
As the samples were processed at different institutions and assayed using different array platforms and protocols, considerable systematic biases were expected to be manifested in the data sets as differences in gene expression patterns. In order to identify and adjust systematic biases imposed by characteristics of the different array platforms, we used the method of DWD.27 Following this procedure, there was a clear improvement in the correlations of relative expression measurements (Table 4). As before, correlations obtained after normalization by MAS5 were lower than the respective values generated with vsn or gcRMA, unless low-intensity signals were excluded from the analyses. The respective orthogonal regression lines showed little if any change in slope when the data from both platforms had been normalized by the vsn algorithm. Moderate changes were detected upon DWD in case gcRMA had been used to normalize Affymetrix data, whereas normalization by MAS5 tended to cause more severe variation. On average, the slopes were closest to 1 when vsn was used to normalize the Affymetrix data and deviated the most from 1 upon normalization by MAS5. As expected, systematic bias correction by DWD shifted the slopes towards one in almost all cases (Table 4).
Significant Differences and Similarities
A different approach to detect differences between the two array systems is to compare the sets of genes identified as differentially expressed. In Figure 3, volcano plots26 show the log2-ratios of those 2861 genes consistently detected in the four primary HNSCC vs normal mucosa experiments and their respective P-values. The two platforms identified similar numbers of differentially expressed genes (≥2-fold difference), both regarding raw P-values (P≤0.001) or FDR-adjusted P-values (adj. P≤0.1).25 There were 45 genes identified as differentially expressed in all tumor samples on the Affymetrix platform, 53 were scored on Operon arrays, and the intersection contained 21 genes discovered on both systems (Table 5, Figure 4a). Plotting of the corresponding mean log ratios (Figure 4b) revealed that even genes scored by only one of the systems generally showed the same direction, but not the same degree of differential expression on the other. GO data mining29 for ‘biological process’ (at level 3) assigned the majority of annotated genes from each platform to cell growth and/or maintenance as well as various metabolic pathways (Figure 5). However, using the software EASE,30 which performs a statistical analysis of the GO categories assigned to the differentially expressed genes, accounting for the distribution of GO categories in the list of all analyzed genes to find those categories that are the most overrepresented (and can therefore be described as ‘themes’), revealed a trend towards components of the extracellular matrix for both of the platforms. Furthermore, genes involved in lipid metabolism were significantly overrepresented only among the differentially expressed genes identified on the Affymetrix system, whereas the Operon platform additionally detected genes engaged in ion binding (Table 6).
Volcano plots of P-values against log2-transformed expression ratios. Mean log2-ratios of tumor vs reference samples are shown on the x-axis. The corresponding P-values of significance, derived by empirical Bayes inference (a, b) or empirical Bayes inference and subsequent adjustment to control the FDR (c, d), are displayed on the y-axis. Results are shown for those 2861 genes consistently detected in the four primary HNSCC vs normal mucosa experiments. The plots were segmented to illustrate the relation of statistical significance (P≤0.005, adj. P≤0.1) to significance based on a two-fold change criterion. Only genes indicated by spots in the upper left and right segments of the plots satisfy both criteria, their numbers explicitly shown. Genes located in the lower left and right segments display a large fold-change but fail to achieve statistical significance. Genes found in the middle segments show no relevant difference of expression, with (upper segments) or without (lower segments) additional statistical significance associated with this observation.
Summary of genes scored as differentially expressed with either one or both evaluated platforms. (a) Venn diagram showing subsets of genes that exhibit a significant differential expression with either technology, taken from a pool that contained only those genes that could repeatedly be quantified in the four primary HNSCC vs normal mucosa experiments. (b) log–log plot illustrating the relationship of the log-ratios for the 77 genes shown in (a).
GO data mining. The 45 regulated genes detected with the Affymetrix system as well as the 52 regulated genes found with Operon arrays were characterized according to their biological process classification in the GO database (at level 3). Roughly half of the genes did not have a GO classification at this level. The majority of the remaining genes were involved with cell growth and/or maintenance as well as various metabolic pathways.
RQ-PCR Analysis
For a small subset of genes, we verified differential expression measurements by RQ-PCR analysis (Figure 6). There was good qualitative agreement between the values determined by either GeneChip arrays, Operon arrays or RQ-PCR. All platforms showed the same direction of regulated gene expression. However, the magnitude of differential expression differed considerably depending on both the experimental approach and the algorithm applied for normalization. Firstly, GeneChip intensity measurements were transformed by variance stabilization (vsn), which was also used for the long oligonucleotide arrays and derives an approximately constant variance along the complete intensity range.17 Normalization was additionally accomplished employing the MAS5 algorithm from the current version of the Affymetrix Microarray Suite software package.19 At least for the small number of genes and patients shown here, there is a tendency for higher ratios with vsn normalization.
Comparison of relative gene expression for the genes OSF2, GMDS, TMPRSS2 and BGN. Expression ratios were determined for tumor vs control tissue of the indicated patients, using either Affymetrix GeneChip arrays (a, b), Operon long oligonucleotide arrays (c) or real-time quantitative PCR analysis (d). Affymetrix ratios were either normalized by variance stabilization (a) or the MAS5 algorithm (b).
Discussion
The comparison of gene expression measurements obtained with different technical approaches or different implementations of a proven technology is of considerable interest to researchers from all fields of the biological and biomedical sciences. Several studies have addressed this topic, with rather heterogeneous results.
Recently, Mah et al8 compared absolute expression levels quantified on Affymetrix short oligonucleotide and radioactively labeled cDNA-based filter arrays. The expression values from the two technologies showed merely poor correlations. Tan et al34 evaluated the performance of three commercial microarray platforms and found only modest correlations when comparing both absolute and relative gene expression measurements. Strikingly, log2-ratios from the two platforms using short oligonucleotide probes and biotinylated cRNA targets (Affymetrix and Amersham; r=0.52) did not correlate better with each other than with those of cDNA arrays (Agilent; r=0.53 or r=0.59). In a comparison of Affymetrix GeneChip arrays and two different collections of 70-mer oligonucleotides, Barczak et al10 found moderate correlations of corresponding signal intensities (r=0.56–0.60), but strong correlations of respective relative expression values (r=0.80 without filtering, r=0.83–0.89 after exclusion of probes or probe sets with low signal intensities). Similarly, Shippy et al35 described improved correlations between expression measurements from Affymetrix GeneChip and Amersham CodeLink arrays upon removal of genes within platform noise (r=0.62 vs r=0.79). Measuring relative gene expression values on Affymetrix short oligonucleotide arrays, commercial (Agilent) and custom-made, sequence-validated cDNA arrays, Järvinen et al36 observed reasonable correlations of log2-ratios. Interestingly, the correlation between the two different cDNA platforms (r=0.73) was weaker than the correlations between the commercial or custom-made cDNA arrays and the Affymetrix system (r=0.84 and 0.76, respectively). A recent study by Tan et al,34 showing very poor correlation between Affymetrix, Amersham and Agilent arrays, came to broad public attention37 and raised general concerns regarding the comparability of expression data across labs and platforms.
A different approach to review the possibility for meaningful translation of microarray data is meta-analysis of extensive data sets of similar type, produced in different labs and on different platforms.38, 39, 40 Since many additional parameters such as classification of the samples or individual laboratory practices influence the outcome of these studies, the results are rather inconclusive concerning comparability on the technological level. Generally, at least common patterns and/or groups of genes could be confirmed.
Aside from meta-analyses, the above-mentioned studies were usually based on data generated with homogeneous cell lines and by averaging over several technical replicates. We intended to increase the practical significance of our evaluation by the use of clinical samples in combination with modest technical replication (two single arrays per patient for the Affymetrix platform and two dye swap replicates per patient for the spotted oligonucleotide arrays). Additionally, we kept the protocols for target preparation as comparable as possible. Since the Affymetrix platform utilizes biotinylated cRNA generated by in vitro transcription (IVT), we decided to employ a linear, IVT-based amplification and labelling protocol for the spotted oligonucleotide arrays instead of the usual dye-labelling by reverse transcription. Recently, we showed that this novel procedure generates highly reproducible expression profiles with down to 2 ng of starting material.12 We could also demonstrate that the correlation of expression ratios obtained with spotted oligonucleotide arrays is higher between replicate amplified sample pairs than between amplified and RT-labelled sample pairs or replicate RT-labelled sample pairs. Accordingly, one can expect that consistent target amplification would also be beneficial, if expression ratios are to be compared across platforms. Comparative studies that do not account for this consideration might introduce additional systematic bias, resulting in reduced agreement between platforms.
To match the probes from the two platforms, we used accession numbers provided by Affymetrix and Operon and mapped them to the current version of the UniGene database. Although provided by both manufacturers, transcript identifiers from the RefSeq collection41, 42 were not chosen for matching the platforms, since reference sequences can change through consolidation of the database. Recently, Mecham et al43 showed that up to 50% of Affymetrix probes do not have a matching sequence in the current version of RefSeq. Despite these considerations, platform matching by RefSeq identifiers yielded approximately similar and partly even improved results in terms of cross-platform correlation (9922 genes could be assigned as represented on both platforms, correlations of unfiltered log2-ratios were r=0.66 - r=0.81; data not shown). Evolution of the UniGene database (accession numbers that were removed due to misalignment or retraction by their submitters; UniGene clusters that were retired as they could be joined or split) and the associated loss of cross-references may also explain why we identified less genes common to both array types than previously reported by Barczak et al.10 We also decided against matching by GenBank accession numbers, since corresponding probes and probe sets can be annotated by different accession numbers of the same UniGene, causing this procedure to exclude large amounts of potentially useful information.
When comparing unfiltered, log2-transformed expression ratios of individual patients, obtained with either GeneChip short oligonucleotide arrays or spotted long oligonucleotide arrays, we detected considerable variations in the degree of correlation (Figure 2). As reported previously,10, 44 these correlations improved after the exclusion of probes and probe sets associated with low signal intensities. This observation might, at least in part, be attributed to variations in the performance of individual array experiments.
Systematic bias adjustment by DWD could further improve the correlations between expression ratios (Table 4). DWD is an advanced method for the adjustment of various systematic differences across microarray experiment subpopulations, including sample source, batch and platform effects,27 which facilitates the merging of different data sets. DWD uses an approach similar to that of support vector machines (SVM),45 but delivers improved performance in the context of high-dimensional, low sample size (HDLSS) data such as those obtained by microarray analyses. Both methods aim at finding a hyperplane in high-dimensional space, which separates defined subpopulations of data as completely as possible. The essential difference is that, while SVM tries to maximize the minimum distance (margin) of all the data to the separating plane, DWD works by maximizing the sum of the inverse distances. In this way, all data points have an influence on the result (optimized position of the hyperplane), and data piling at the margins is avoided, a problem associated with the minimum distance criterion of SVM. After determination of the DWD direction vector, all data points of each subpopulation are projected onto the direction given by this vector. Finally, data points from each subpopulation are shifted in the DWD direction by subtracting the DWD direction vector multiplied by their projected means, thereby effectively removing systematic variation while preserving any variation in the DWD direction not caused by systematic effects. Applied to our data, the DWD approach clearly and consistently improved cross-platform correlations while shifting the slopes of corresponding regression lines towards 1 (Table 4). The latter effect was minimal in case both data sets had been normalized by the same algorithm (vsn), as this procedure not unexpectedly yielded slopes closest to one even before DWD. A slope close to one implies that genes are more likely to yield similar results (regardless of differential expression) on both of the investigated platforms. Further improvements of DWD performance can be anticipated for more extensive data sets, and we are confident that this method could greatly enhance agreement in future comparative studies.
Our goal was to compare reliable measurements from both systems, both of which can be regarded as detecting overlapping but different subsets of the actual set of differentially expressed genes. This was confirmed by EASE overrepresentation analysis,30 which revealed that some of the differentially expressed genes could be assigned to the same ‘theme’ on both platforms, whereas others were exclusive to one of the platforms (Table 6). On each array system, approximately 50 genes were consistently and repeatedly scored as differentially expressed, and the intersection of these groups contained 21 common genes (Figure 4a). The majority of genes restricted to one of the platforms showed no sufficient degree and/or significance, but at least the same direction of regulated expression on the other platform (Figure 4b). Therefore, it does not matter if a clinical study uses Affymetrix or Operon long oligonucleotide arrays, as long as these are used consistently and combined with high quality control standards throughout the whole investigation.
For a subset of genes, we verified microarray-derived expression ratios by RQ-PCR and found good qualitative agreement between the two array platforms and the PCR-based method (Figure 6).
We have shown that, overall, expression profiles obtained with either long (Operon) or multiple short (Affymetrix) oligonucleotide microarrays display a reasonable correlation, with variable concordance of individual genes. Based on patient samples, we obtained results that are in good agreement with previous studies that utilized cell line-derived RNA. Projecting these findings to a larger series of array experiments, one could expect to obtain similar albeit not identical results, concerning, for example, a hierarchical clustering or a gene expression signature, with either of the two investigated platforms. On the level of individual genes and quantitative precision, however, our results reaffirm that microarrays have to be considered a screening technology and that their data should be regarded with caution. This should be kept in mind particularly when comparing data from different array platforms. Recently, important progress has been made to facilitate this transfer of information. Guidelines provided by the ‘Microarray Gene Expression Data Society (MGED)’ (http://www.mged.org), which developed the ‘Minimum Information About a Microarray Experiment (MIAME)’ specifications,46 assist researchers in the annotation of their microarray experiments. Further improvement is provided by public microarray repositories, which facilitate the publication and sharing of properly annotated gene expression data. Statistical methods like DWD27 can further improve the comparability of microarray data sets, since systematic biases arising from platform-specific parameters, such as measurement precision (reproducibility), accuracy (regarding the ‘true’ values), specificity and sensitivity or differences in protocol performance, can be properly weighted and adjusted accordingly. The utility of future array studies could further improve if the ‘External RNA Control Consortium (ERCC)’ is successful in its effort to standardize controls for the calibration of microarray experiments. But ultimately, meaningful comparison, translation and integration of expression data will be impaired as long as industrial standards are missing for the production of arrays as well as for the design of array probes.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995;270:467–470.
Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 1996;14:1675–1680.
Hughes TR, Mao M, Jones AR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol 2001;19:342–347.
Wrobel G, Schlingemann J, Hummerich L, et al. Optimization of high-density cDNA-microarray protocols by ‘design of experiments’. Nucleic Acids Res 2003;31:e67.
Kokocinski F, Wrobel G, Hahn M, et al. QuickLIMS: facilitating the data management for DNA-microarray fabrication. Bioinformatics 2003;19:283–284.
Knight J . When the chips are down. Nature 2001;410:860–861.
Yuen T, Wurmbach E, Pfeffer RL, et al. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res 2002;30:e48.
Mah N, Thelin A, Lu T, et al. A comparison of oligonucleotide and cDNA-based microarray systems. Physiol Genomics 2004;16:361–370.
Schlingemann J, Hess J, Wrobel G, et al. Profile of gene expression induced by the tumour promotor TPA in murine epithelial cells. Int J Cancer 2003;104:699–708.
Barczak A, Rodriguez MW, Hanspers K, et al. Spotted long oligonucleotide arrays for human gene expression analysis. Genome Res 2003;13:1775–1785.
Cox WG, Beaudet MP, Agnew JY, et al. Possible sources of dye-related signal correlation bias in two-color DNA microarray assays. Anal Biochem 2004;331:243–254.
Schlingemann J, Thuerigen O, Ittrich C, et al. Effective transcriptome amplification for expression profiling on sense-oriented oligonucleotide microarrays. Nucleic Acids Res 2005;33:e29.
UICC. TNM Classification of Malignant Tumours, 6th edn. John Wiley & Sons: New York, 2002.
Schena M, Shalon D, Heller R, et al. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA 1996;93:10614–10619.
R Development Core Team. R: A Language and Environment for Statistical Computing, 2nd edn. R Foundation for Statistical Computing: Vienna, 2004.
Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80.
Huber W, Von Heydebreck A, Sultmann H, et al. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002;18 Suppl. 1:S96–S104.
Affymetrix. GeneChip Expression Analysis Technical Manual, 5th edn. Affymetrix: Santa Clara, 2004.
Affymetrix. Microarray Suite User Guide, 5th edn. Affymetrix: Santa Clara, 2001.
Wu Z, Irizarry R, Gentleman R, et al. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 2004;99:909–917.
Zhang J, Carey V, Gentleman R . An extensible application for assembling annotation for genomic data. Bioinformatics 2003;19:155–156.
Yang MC, Ruan QG, Yang JJ, et al. A statistical method for flagging weak spots improves normalization and ratio estimates in microarrays. Physiol Genomics 2001;7:45–53.
Smyth GK, Yang YH, Speed T . Statistical issues in cDNA microarray data analysis. Methods Mol Biol 2003;224:111–136.
Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article 3.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300.
Wolfinger RD, Gibson G, Wolfinger ED, et al. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 2001;8:625–637.
Benito M, Parker J, Du Q, et al. Adjustment of systematic microarray data biases. Bioinformatics 2004;20:105–114.
Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–29.
Dennis Jr G, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:P3.
Hosack DA, Dennis Jr G, Sherman BT, et al. Identifying biological themes within lists of genes with EASE. Genome Biol 2003;4:R70.
Pfaffl MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45.
Edgar R, Domrachev M, Lash AE . Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210.
Baugh LR, Hill AA, Brown EL, et al. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res 2001;29:E29.
Tan PK, Downey TJ, Spitznagel Jr EL, et al. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res 2003;31:5676–5684.
Shippy R, Sendera TJ, Lockner R, et al. Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations. BMC Genomics 2004;5:61.
Jarvinen AK, Hautaniemi S, Edgren H, et al. Are data from different gene expression microarray platforms comparable? Genomics 2004;83:1164–1168.
Marshall E . Getting the noise out of gene arrays. Science 2004;306:630–631.
Rhodes DR, Barrette TR, Rubin MA, et al. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res 2002;62:4427–4433.
Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003;100:8418–8423.
Mitchell SA, Brown KM, Henry MM, et al. Inter-platform comparability of microarrays in acute lymphoblastic leukemia. BMC Genomics 2004;5:71.
Pruitt KD, Katz KS, Sicotte H, et al. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet 2000;16:44–47.
Pruitt KD, Maglott DR . RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res 2001;29:137–140.
Mecham BH, Wetmore DZ, Szallasi Z, et al. Increased measurement accuracy for sequence-verified microarray probes. Physiol Genomics 2004;18:308–315.
Baum M, Bielau S, Rittner N, et al. Validation of a novel, fully integrated and flexible microarray benchtop facility for gene expression profiling. Nucleic Acids Res 2003;31:e151.
Boser BE, Guyon IM, Vapnik VN, Proceedings of the Fifth Annual Workshop on Computational Learning Theory. ACM Press: Pittsburgh, 1992, pp 144–152.
Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 2001;29:365–371.
Acknowledgements
We thank Axel Benner for helpful discussions and critical reading of the manuscript as well as Bernd Groner for excellent support. Bjoern Tews and Silke Deckert are gratefully acknowledged for their skillful technical assistance. Our study was supported by grants of the Bundesministerium für Bildung und Forschung (FKZ KR-S07T13 to RS; NGFN-1, 01 GR 0101 to PL; NGFN-2, 01 GS 0460 and 01 GR 0418 to PL and MH) as well as the EU MolTools project (LSHG-CT-2004-503155 to PL).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schlingemann, J., Habtemichael, N., Ittrich, C. et al. Patient-based cross-platform comparison of oligonucleotide microarray expression profiles. Lab Invest 85, 1024–1039 (2005). https://doi.org/10.1038/labinvest.3700293
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700293
Keywords
This article is cited by
-
Integrated computational analysis reveals HOX genes cluster as oncogenic drivers in head and neck squamous cell carcinoma
Scientific Reports (2022)
-
Clinical correlation of opposing molecular signatures in head and neck squamous cell carcinoma
BMC Cancer (2019)
-
Nanosized food additives impact beneficial and pathogenic bacteria in the human gut: a simulated gastrointestinal study
npj Science of Food (2018)
-
Expressional analysis of disease-relevant signalling-pathways in primary tumours and metastasis of head and neck cancers
Scientific Reports (2018)
-
The actin-myosin regulatory MRCK kinases: regulation, biological functions and associations with human cancer
Journal of Molecular Medicine (2014)